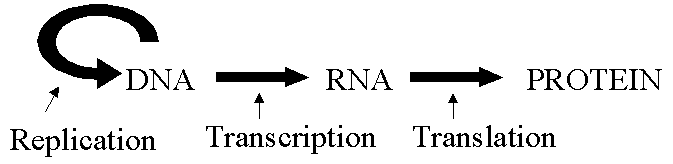
Lecture 1. Genetics is a science of genes
Since the beginning of human history, people have wondered how traits are inherited from one generation to the next. Although children often look more like one parent than the other, most offspring seem to be a blend of the characteristics of both parents. Centuries of breeding of domestic plants and animals had shown that useful traits - speed in horses, strength in oxen, and larger fruits in crops - can be accentuated by controlled mating. However, there was no scientific way to predict the outcome of a cross between two particular parents.
It wasn't until 1865 that an Augustinian monk named Gregor Mendel found that individual traits are determined by discrete "factors," later known as genes, which are inherited from the parents. His rigorous approach transformed agricultural breeding from an art to a science. However, Mendel’s work was not appreciated immediately.
That’s why the science of genetics really began with the rediscovery of Gregor Mendel's work at the turn of the 20th century, and the next 40 years or so saw the elucidation of the principles of inheritance and genetic mapping. Microbial genetics emerged in the mid 1940s, and the role of DNA as the genetic material was firmly established. During this period great advances were made in understanding the mechanisms of gene transfer between bacteria, and a broad knowledge base was established from which later developments would emerge.
The discovery of the structure of DNA by James Watson and Francis Crick in 1953 provided the stimulus for the development of genetics at the molecular level, and the next few years saw a period of intense activity and excitement as the main features of the gene and its expression were determined. This work culminated with the establishment of the complete genetic code in 1966. The stage was now set for the appearance of the new genetics.
From 1865 to now the history of genetics development is the development of human knowledge and understanding of genes. In other words, genetics is a science of the structure, function and movement of genes. Before going into the exact definition of gene, one can begin by understanding that a gene is a piece of DNA which has a function such as determining human eye color, pea seed shape or a disease.
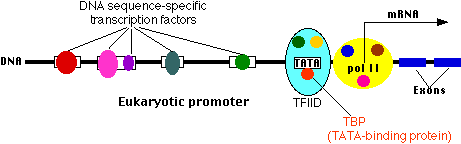
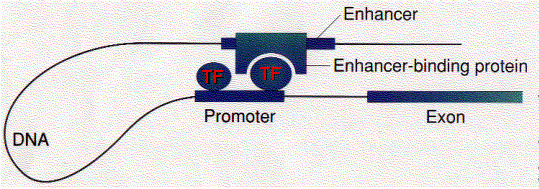
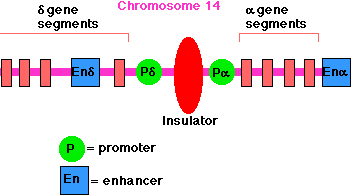
Lecture 2. Genes are mostly located on chromosomes
All living organisms are composed of cells. Many of the chemical reactions of an organism, its metabolism, take place inside of cells. The genetic information required for the maintenance of existing cells and the production of new cells is stored within the membrane-bound nucleus in eukaryotic cells or in the nucleoid region of prokaryotes. This genetic information passes from one generation to the next.
The nucleus, which contains the genetic information (DNA), is the control center of the cell. DNA in the nucleus is packaged into chromosomes. DNA replication and RNA transcription of DNA occur in the nucleus. Transcription is the first step in the expression of genetic information and is the major metabolic activity of the nucleus.
A gene, a unit of hereditary information, is a stretch of DNA sequence, encoding information in a four-letter language in which each letter represents one of the nucleotide bases. Much of the information stored in stretches of DNA sequence is subsequently expressed as another class of biopolymers, the proteins.
Work on cytology in the late 1800s had shown that each living thing has a characteristic set of chromosomes in the nucleus of each cell. During the same period, biochemical studies indicated that the nuclear materials that make up the chromosomes are composed of DNA and proteins. In the first four decades of the 20th century, many scientists believed that protein carried the genetic code, and DNA was merely a supporting "scaffold." Just the opposite proved to be true. Work by Avery and Hershey, in the 1940s and 1950s, proved that DNA is the genetic molecule.
Work done in the 1960s and 1970s showed that each chromosome is essentially a package for one very long, continuous strand of the DNA. In higher organisms, structural proteins, some of which are histones, provide a scaffold upon which DNA is built into a compact chromosome. The DNA strand is wound around histone cores, which, in turn, are looped and fixed to specific regions of the chromosome.
Lecture 3. Genes are made of DNA or RNA
Structure of DNA
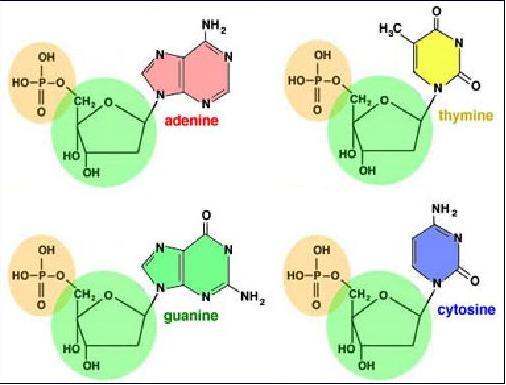
Deoxyribonucleic acid (DNA) is composed of building blocks called nucleotides consisting of a deoxyribose sugar, a phosphate group, and one of four nitrogen bases - adenine (A), thymine (T), guanine (G), and cytosine (C). Phosphates and sugars of adjacent nucleotides link to form a long polymer. It was showed that the ratios of A - to T and G – to - C are constant in all living things. X-ray crystallography provided the final clue that the DNA molecule is a double helix, shaped like a twisted ladder.
In 1953, the race to determine how these pieces fit together in a three-dimensional structure was won by James Watson and Francis Crick at the Cavendish Laboratory in Cambridge, England. They showed that alternating deoxyribose and phosphate molecules form the twisted uprights of the DNA ladder. The rungs of the ladder are formed by complementary pairs of nitrogen bases - A always paired with T and G always paired with C.
Base pairs bond the double helix together. The "beginning" of a strand of a DNA molecule is definedas 5'. The "end" of the strand of A DNA molecule is defined as 3'. The 5' and 3' terms refer to the position of the nucleotide base, relative to the sugar molecule in the DNA backbone, which is make up by the phosphodiester bonds linking between the 3' carbonatom and the 5' carbon of the sugar deoxyribose (in DNA) or ribose (in RNA).
Each chromosome is composed of a single DNA molecule. Our DNA contains greater than 3 billion base pairs--an enormous amount by any measure. All of this information must be organized in such a manner that it can be packaged inside the nucleus of the cell. To accomplish this, DNA is complexed with histones to form chromatin. Histones are special proteins that the DNA molecule coils around to become more condensed. The chromatin then becomes coiled upon itself, which ultimately forms chromosomes.
When one cell divides into two daughter cells, the DNA, all 46 chromosomes, for example, in humans, must be replicated. The specificity of base pairing between A/T and C/G is essential for the synthesis of new DNA strands that are identical to the parental DNA. Each strand of DNA serves as a template for DNA synthesis. Synthesis occurs by adding bases that exactly mirror the template strand. So, as each strand is copied, two sets of DNA are made that are identical to the original two strands. The order of nucleotide bases along a DNA strand is known as the sequence.
If a problem occurs during DNA replication, this can lead to a disruption of gene function. For example, if the wrong base is inserted during replication (a mutation) and this mistake happens to be in the middle of an important gene, it could result in a non-functional protein. Fortunately, we have evolved various mechanisms to ensure that such mutations are detected, repaired, and not propagated. However, these mechanisms sometimes fail, and uncorrected mutations will occur. If the resulting alteration in gene function, through its interplay with the environment, sufficiently disrupts metabolism or structure, clinical disease can result.
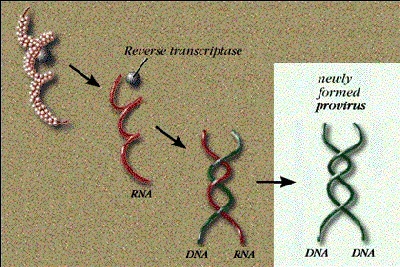
Some viruses store genetic information in RNA
DNA was believed to be the sole medium for genetic information storage. Furthermore, Watson and Crick's central dogma assumed that information flowed "one-way" from DNA to RNA to protein. So it came as a surprise in 1971 when it was discovered that some viruses’ genetic information is RNA.
Even so, these viruses ultimately make proteins in the same way as higher organisms. During infection, the RNA code is first transcribed "back" to DNA - then to RNA to protein, according to the accepted scheme. The initial conversion of RNA to DNA - going in reverse of the central dogma - is called reverse transcription, and viruses that use this mechanism are classified as retroviruses. A specialized polymerase, reverse transcriptase, uses the RNA as a template to synthesize a complementary and double stranded DNA molecule as shown in the picture.
Lecture 4. Genes can replicate themselves
As genes are made of DNA, they can make themselves when DNA is replicated. The specificity of base pairing between A/T and C/G helps explain how DNA is replicated prior to cell division. Enzymes unzip the DNA by breaking the hydrogen bonds between the base pairs. The unpaired bases are now free to bind with other nucleotides with the appropriate complementary bases. The enzyme primase begins the process by synthesizing short primers of RNA nucleotides complementary to the unpaired DNA. DNA polymerase now attaches DNA nucleotides to one end of the growing complementary strand of nucleotides. Replication proceeds continuously along one strand, called the leading strand, which is shown here on the right. The process occurs in separate short segments called Okazaki fragments next to the other, or lagging, strand on the left. This difference is due to the fact that DNA polymerase can only add new nucleotides to the 3 prime end of a nucleotide strand in a 5’ 3’ direction. A primer begins any new strand, including each Okazaki fragment. An enzyme replaces the RNA primer with DNA nucleotides. Then an enzyme called DNA ligase binds the fragments to one another.
There are now two DNA molecules. Each consists of an original nucleotide strand next to a new complementary strand. The two molecules are identical to each other.
A detailed and clear schematic of DNA synthesis kindly provided by Prof. Douglas J. Burks is shown below:
http://upload.wikimedia.org/wikipedia/commons/thumb/9/9f/DNA_replication.svg/691px-DNA_replication.svg.png
Lecture 5. Language of genes is simple and informative
Genetic information likes a language. We use letters of the alphabet to make words and then join these words together to make sentences, paragraphs and books. In the case of DNA:
The alphabet is only 4 letters (A,T,G and C) long.
Each letter represents a chemical compound called a base or nucleotide .
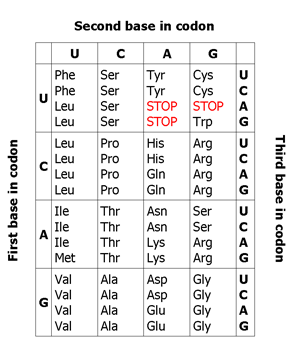
These 4 letters are used to form the genetic words called codons.
Unlike a normal language, all genetic words are only three letters long.
These words combine together to form sentences called genes, which encode the instruction for amino acids in a polypeptide.
At the end of each sentence is a special word or full stop called a stop codon.
All the sentences join together to form a book that contains all the genetic information about you called your genome.
Let’s make some comparisons between English Language and Genetic Language:
Table 2.1. | English Language
FIXME: A LIST CAN NOT BE A TABLE ENTRY.
We use 26 letters to make words. The words can be any length we need. We join words together to create sentences Each sentence starts with a capital letter.Each sentence ends with a fullstop.All the sentences combine to form a book. | Genetic Language
FIXME: A LIST CAN NOT BE A TABLE ENTRY.
DNA uses 4 molecules to make codons. The codons can only be 3 nucleotides long. The codons join together to form genes. The gene starts with codon AUG.The gene stops at a specific stop codon. All the genes combine to form the genome. |
The Genetic Language of DNA provides the information needed to produce proteins. It is these proteins that carry out the biochemical processes (metabolism) to ensure an organism's ongoing survival. Proteins have their own language that has an "alphabet" of 20 "letters". These letters are the amino acids. RNA is used to "translate" genetic language into protein language. It takes the information from a gene of the DNA strand and creates the proteins necessary for life.
Along the gene (and DNA itself) the information for the amino acids that will make up the gene is stored in three-letter words called codons. Each codon specifies a particular amino acid. By "reading" this set of codons, the specific protein can be generated from this chunk of genetic code. The codons on DNA code for a specific amino acid. There are 20 amino acids commonly found in natural proteins.
Below is a “paragraph” of gene language:
CCG ACG TCC GAA GAG TGA CCG ACG TCC GAA GAG TGA CCG ACG TCC GAA GAG TGA CCG ACG TCC GAA GAG TGA CCG ACG TCC GAA GAG TGA CCG ACG TCC GAA GAG TGA CCG ACG TCC GAA GAG TGA CCG ACG TCC GAA GAG TGA CCG ACG TCC GAA GAG TGA CCG ACG TCC GAA GAG TGA CCG ACG TCC GAA GAG TGA CCG ACG TCC GAA GAG TGA CCG ACG TCC GAA GAG TGA CCG ACG TCC GAA GAG GAA GAG TGA CCG ACG TCC GAA GAG TGA CCG ACG TCC GAA GAG TGA CCG ACG TCC GAA GAG TGA CCG
Lecture 6. Altered genes are mutations
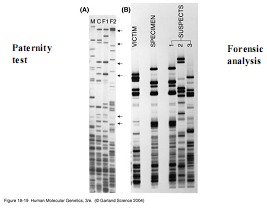
The DNA sequences from two individuals of the same species are highly similar - differing by only about one nucleotide in 1,000. A mutation is, most simply, an alteration in a DNA sequence. This change may or may not lead to a change in the protein coded by the gene. A change that has no effect on protein sequence or function is termed a polymorphism and is a part of the normal variation present in the human genome. Often, however, a change in a DNA sequence will result in the disruption of gene function that we term "Clinical Manifestations" in the Clinical Integration Model. The altered protein that results from a mutation can disrupt the way a gene functions, and this can lead to clinical disease. How these mutations manifest themselves depends on each individual's unique genetic endowment and interactions with their environment.
Furthermore, the change may or may not be passed on to subsequent generations. If, as in non-familial cancer, the mutation occurs in isolated somatic cells, it will not be passed on to subsequent generations. Only those mutations occurring to the DNA in the gametes (egg or sperm) will potentially be passed on to offspring. If the mutation is passed on to the offspring, they will carry this mutation in all of the cells in their body.
Following is a brief review of different types of mutations:
Base pair substitution
Replacement of one DNA base by another in the DNA sequence. Replacement of nucleotide bases can have several possible consequences.
Missense mutation
An amino acid residue in the original protein may be replaced by a different one in the mutated protein.
Nonsense mutation
The codon for an amino acid residue within the original protein is changed to a stop codon, which leads to a premature termination of the protein resulting in a non functional protein.
Silent Mutation
The codon for an amino acid is changed, but the same amino acid is still coded for. This is possible because some amino acids are coded for by multiple codons. For example, the sequences UGC and UGU both code for Cysteine.
Frameshift mutation
A deletion or insertion of any number of bases other than a multiple of three bases has a much more profound effect. Such frameshift mutation results in a complete change in the amino acid sequence downstream from the point of mutation, instead of simply a change in the number of amino acids.
Deletions, Insertions, and Duplications
Deletions or insertions may be large or small. Large insertions and deletions in coding regions almost invariably prevent the production of useful proteins. The effect of short deletions or insertions depends on whether or not they involve multiples of three bases. If one, two, or more whole codons (three base pairs or any multiple of three) are removed or added, the consequence is the deletion or addition of a corresponding number of amino acid residues. Sometimes, an entire gene can be inserted (duplicated) or deleted. The effects of these types of mutations depend on where in the genome they occur and how many base pairs are involved.
Normal
THE BIG RED DOG RAN OUT.
Missense
THE BIG RAD DOG RAN OUT.
Nonsense
THE BIG RED.
Frameshift - deletion
THE BRE DDO GRA.
Frameshift - insertion
THE BIG RED ZDO GRA.
Inversions
This type of mutation occurs when a chromosomal section is separated from the chromosome, rotates 180 degrees, and rejoins the chromosome in an opposite orientation. This type of mutation can affect a gene at many levels. If an inversion disrupts a promoter region, the gene may not be transcribed at all. If the coding sequence is disrupted, a non-functional gene product (protein) may result.
Translocations
This type of chromosomal aberration results when one portion of a chromosome is transferred to another chromosome. This can be a very harmful event if it leads to a subsequent gain or loss of genetic material. Additionally, when a gene from one chromosome moves to another chromosome, large changes in the ability to regulate expression of the gene may occur. Some forms of leukemia result from translocations. In these cases, various genes controlling growth of white blood cells are constantly turned on, leading to an uncontrolled proliferation of these cells and the various clinical manifestations of leukemia.
LacZ mutations*
LacZ mutations are an example of particular mutations found in the LacZ gene of E.coli, which encodes the lactose hydrolyzing enzyme ß-galactosidase. There is a special compound known as X-gal that can be hydrolyzed by ß-galactosidase to release a dark blue pigment. When X-gal is added to the growth medium in petri plates, Lac+ E. coli colonies turn blue, whereas Lac– colonies with mutations in the LacZ gene are white. By screening many colonies on such plates it is possible to isolate a collection of E. coli mutants with alterations in the LacZ gene. PCR amplification of the LacZ gene from each mutant followed by DNA sequencing allows the base changes that cause the LacZ– phenotype to be determined. A very large number of different LacZ mutations can be found, but they can be categorized into three general types: missense, nonsense and frameshift .
Causes of mutations
Mutations are caused by substances that disrupt the chemical structure of DNA or the sequence of its bases. Radiation, various chemicals, and chromosome rearrangements are some of the many sources of mutation.
Mutation rates
All of us are subjected to mutagenic events throughout our lifetime. Depending upon the type of mutation, the frequency ranges from 10-2/cell division to 10-10/cell divisio