CHAPTER 3
METHODOLOGY
3.1 Sampling :
Intravenous blood samples were collected from a total of 100 healthy
unrelated individuals belonging to Bhumij tribe , which are Austro-Asiatic
groups.Vacutainers were used to store blood with ice gels to maintain the
cold temperature. Vacutainers contains EDTA (the potassium salt, or
K2EDTA). This is a strong anticoagulant and these tubes are usually used
for full blood counts (CBC) and blood films.Blood can be stored in it for
45 days from the date of collection.
3.2 MATERIALS & METHODS:
Blood Collection Kit
DISPOVAN® 10ml sterile syringes and VACUETTE® tubes were used for
blood collection. The interior of the tube wall is coated with EDTA K3.
The tube is also available with an 8% liquid EDTA solution. The EDTA
binds calcium ions thus blocking the coagulation cascade. Erythrocytes,
leucocytes and thrombocytes are stable in EDTA anticoagulated blood for
up to 24 hours at 4o C.
Fig 9 : Vacutainer Fig 10 : Transfering blood from
syringe to vacutainer
Materials required :
• Gloves
• Apron
• 50 ml Falcon tubes
• 14 ml Falcon tubes
• Tube stand
• Gel Tray
• Combs
• Electrophoresis tank
• Marker
• Tissue roll
• Autoclave Tape
• Sequencing plates
• Plate flap
• Pipettes
• 1.5 ml eppendorf
• PCR vials
• Ice
• Pippete Tips
• Aluminium foil
• Agrose
• 10 ml Disposable syringe
• Torniquet
• Cotton
• Petriplate
• Flask 250 ml
• Measuring cylinder
• Water bath
Reagents Required:-
REAGENT A (TRITONATE BUFFER ):
Tris HCl (pH 8.0)
-
10ml (pH8)
Sucrose
-
109.54 gm(320mM) for osmoregulation
MgCl2
-
5 ml(5mM) for creation of pore on cell surface
Triton X 100
-
10 ml to lyse the RBCs
DDW - 1000ml (autoclaved)
Reagent B (Lysis Buffer II):
Tris HCl (pH 8.0)
-
40ml(400mM)
NA-EDTA
-
12ml(60mM)
NaCl
-
15ml(150mM)
SDS
-
5ml
REAGENT C:
Sodium per chlorate -
35.115 gm
Milli Q water - 50 ml
TRIS SATURATED ALCOHOL:
Phenol
-
Distilled
8-Hydroxy Quinoline -
0.1%
Tris HCl (pH 8.0)
-
0.5 M
Tris HCl (pH 8.0)
-
0.1 M
CHLOROFORM : ISOAMYL ALCOHOL (24:1):
Chloroform
-
24ml
Isoamyl Alcohol
-
1ml
T. E. BUFFER
Tris HCl (pH 7.5)
-
1ml (10mM)
EDTA (pH 8.0)
-
0.2 ml (1mM)
Make it upto 100ml with DDW
70% ALCOHOL:
Absolute Alcohol
-
70ml
DDW
-
30ml
REAGENTS USED FOR GEL ELECTROPHORESIS:-
1 X TAE BUFFER:
10 x TAE Buffer - 50 ml
DDW - 950 ml
6 X LOADING DYE:
Bromophenol Blue
-
0.125g
Xylene Cyanol FF
-
0.125g
Glycerol
-
15ml
(Diluted with DDW to make up volume to 50ml)
ETHIDIUM BROMIDE:
Ethidium Bromide -
10mg
DDW
-.
1ml
(Stored in dark bottles)
PCR COMPONENTS :
Master Mix (6µl) : + 4 µl DNA
Milli Q – Make the master mix upto 6µl
PCR Buffer - 1 × (no. of samples)n µl
MgCl2 – 0.8 × n µl
Forward Primer : M23 primer – 0.05 × n µl
M12 primer – 0.1 × n µl
M15 primer – 0.14 × n µl
M95 primer – 0.2 × n µl
M82 primer – 0.2 × n µl
Reverse Primer: M23 primer – 0.05 × n µl
M12 primer – 0.1 × n µl
M15 primer – 0.14 × n µl
M95 primer – 0.2 × n µl
M82 primer – 0.2 × n µl
Dntps – 0.6 × n µl
Taq Polymerase - 1 × n µl
REAGENTS FOR PCR SEQUENCING AND PROCESSING:
Big Dye – 25µl
Sequencing Buffer – 175µl
Formamide – 500 µl
Milli Q - 500µl
80% Ethanol – 9600 µl
3M Sodium Acetate – 120µl
Absolute alcohol – 3 ml












INSTRUMENTS USED:
•
Centrifuge (Eppendorff 5810R, Biofuge, Remi R8C)
•
PCR Thermo Cyclers (MJ Research PTC 200, Gene Amp 9700,
Eppendorff, Veriti)
Fig11 : MJ Research PCR












Fig 12 : PCR ( Eppendorf and Veriti)
•
Electrophoresis Apparatus (Pharmacia Biotech EPS600, Hoefer power
pack)
•
Trans illuminator (Syngene)
•
Vortex
•
ABI PRISM® 3730xl DNA Analyzer
The ABI PRISM® 3730xl DNA Sequencer automatically analyzes DNA
molecules labeled with multiple fluorescent dyes. It consists of a charge
couple device (CCD) camera and a power Macintosh® computer that includes
software for data collection and data analysis. After samples are loaded onto








the system’s vertical gel, they undergo electrophoresis, laser detection, and
computer analysis. Electrophoretic separation can be viewed on-screen in real-
time.
Fig 13: DNA Sequencer
SEQUENCE ANALYSIS SOFTWARES:
Sequencing Analysis Software™ Ver. 5.2
Two software packages automatically process gel files or raw sample
files to analyze sample files with base calls matching sequence peaks.
Sequencing Analysis Software™ Ver. 5.2 is used for analysis of data
for 3730 and 3730xl genetic analyzers running on a Mac® OS platform.
Sequencing Analysis Software™ is powered by multiple base caller
algorithms to perform signal processing and classification of peaks
from raw data collected from ABI PRISM® Genetic Analyzers. The
result yields accurate sequence data with electropherograms that can be
viewed by Sequencing Analysis Software™ or Edit View software. If
the KB basecaller is used. It defines and displays mixed bases along
with calculated quality values. It calculates clear range and sample
score. It creates output files in ABI (.seq), FASTA (.seq), Phred
(.phd.1), and standard chromatogram format (.scf) formats. It also
generates an analysis report containing sample analysis statistics.
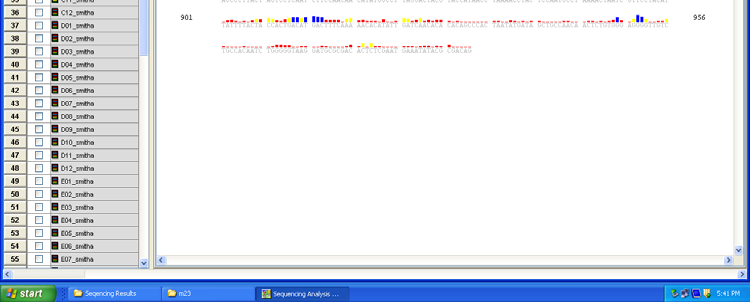
Fig 14: DNA sequencing analysis software

Auto Assembler Version 3.1.2
This is a sequence assembly program and can handle at least 1000 sequences
of 500 bp. It allows on-screen alignment of chromatograms. The manufacturer
claims that the software has no known limitations or bugs. It certainly has the
nice feature of lining up all the electropherograms under each other making
analysis easier. Moreover, it is user-friendly for editing process.
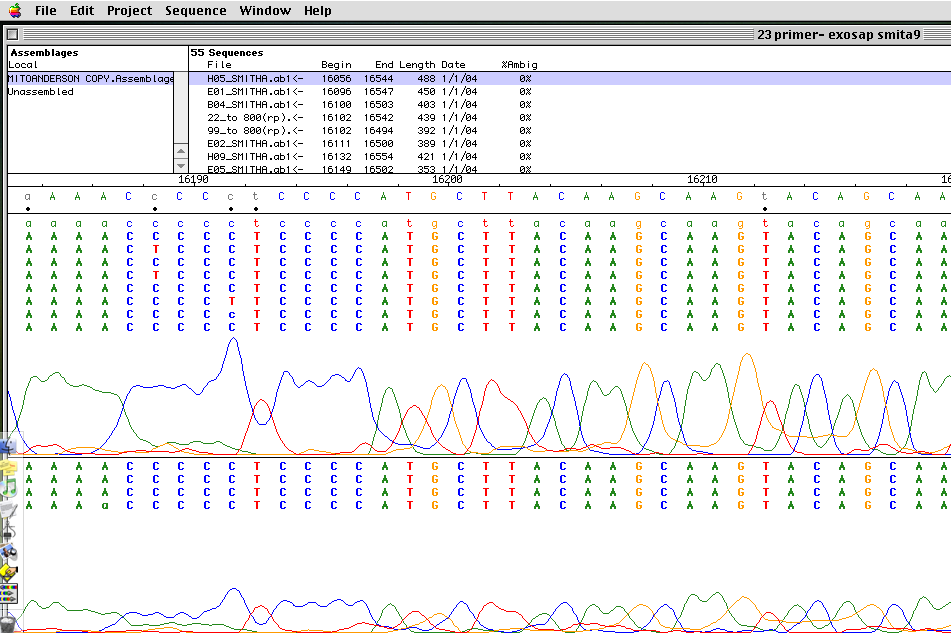
Fig 15: Auto Assembler Software
Protocol :
Isolation of DNA from blood:
DNA was isolated by Phenol-Chloroform method modified by Dr.
Thangaraj (17)(b)(18)
To 9ml of blood sample, 36ml of Reagent-A was added in a 50ml
polypropylene tube. The solution was mixed gently till the solution became
clear.
The above solution was centrifuged at 2,700 rpm for 7min to obtain a
pellet free from RBCs. The supernatant containing lysed RBCs were
discarded carefully.
The pellet was disturbed thoroughly and half the volume as that of
blood sample (roughly 2ml) of Reagent B was added. The solution was
mixed thoroughly by inverting very gently for 3-4min till the solution
became viscous.
To the above solution, 500µl of Reagent C was added and mixed
gently for 3-4min. It precipitates protein molecules which may inhibit PCR.
2ml each of phenol and chloroform was added to the above mixture.It
is a deproteinising reagent.
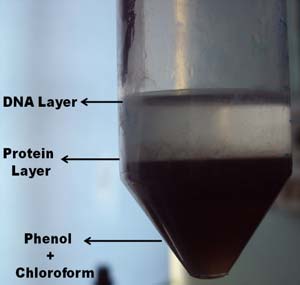
It was mixed well and centrifuged at 3,500 rpm for 8min to separate 3
layers viz. aqueous layer, protein layer and solvent layer.
Fig 16 : aqueous layer, protein layer and solvent layer.

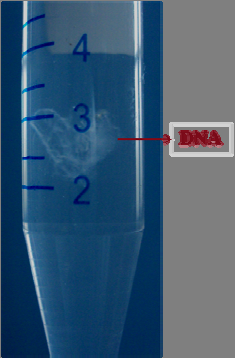


The aqueous layer containing DNA was carefully transferred into a 15ml
polypropylene centrifuge tube using a broad mouth tip.
3ml of chloroform was added to the supernatant and mixed gently for 1 min. It
was then centrifuged at 2,000 rpm for 5min. Chloroform removes left over
phenol and protein which hinders PCR.
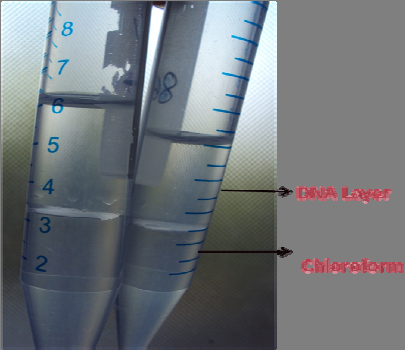
Centrifugation resulted to give 2 clear layers of DNA and Chloroform .
Fig 17 : 2 clear layers of DNA and Chloroform .
The aqueous phase having DNA obtained was transferred to a fresh
polypropylene centrifuge tube.
To this phase, 2ml of chilled isopropyl alcohol was added. It was mixed gently
to precipitate the DNA.
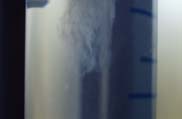
Fig 18 : DNA Extracted
The DNA thread was spooled out and transferred to a fresh Eppendorf tube.
The DNA was washed twice with 70% alcohol and vortexed for 10
seconds.
The pellet was dried properly for 10-15 min to ensure that whole alcohol
had dried off.
The pellet was dissolved in 100 ml of TE Buffer and incubated in water
bath at 55ºC for 45 min to enhance the dissolution.
The DNA samples were stored at 4ºC.
Dilution of DNA:-
5 µl of DNA was mixed in 450 µl of TE buffer in a fresh eppendorf.
It was incubated in 4oC overnight.
Gel Electrophoresis :-
0.96 gm of agrose was added to 120 ml of 1 x TAE buffer.
It was boiled and cooled to room temperature.
1 drop of EtBr was added and gel was casted in the gel tray.
5µl of diluted DNA was loaded with loading dye in the gel.
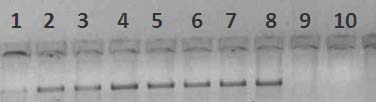
Bands were analysed under UV transilluminator to confirm proper dilution.
Fig 19 : Gel Check of Dilution
PCR :-
4µl diluted DNA was added to labeled autoclaved PCR vials.
Master mix of 6µl was added to each vial.
Master mix for 23rd primer:
1. Milli Q – 2.5 × n (no. of samples)
2. PCR Buffer – 1 × n (no. of samples)
3. MgCl2 – 0.8 × n (no. of samples)
4. Forward primer – 0.05 × n (no. of samples)
5. Reverse primer – 0.05 × n (no. of samples)
6. Dntps – 0.6 × n (no. of samples)
7. Taq polymerase – 1 × n (no. of samples)
Master mix for 15th primer:
1. Milli Q – 2.32 × n (no. of samples)
2. PCR Buffer – 1 × n (no. of samples)
3. MgCl2 – 0.8 × n (no. of samples)
4. Forward primer – 0.14 × n (no. of samples)
5. Reverse primer – 0.14 × n (no. of samples)
6. Dntps – 0.6 × n (no. of samples)
7. Taq polymerase – 1 × n (no. of samples)
Master mix for 12th primer:
1. Milli Q – 2.4 × n (no. of samples)
2. PCR Buffer – 1 × n (no. of samples)
3. MgCl2 – 0.8 × n (no. of samples)
4. Forward primer – 0.1 × n (no. of samples)
5. Reverse primer – 0.1 × n (no. of samples)
6. Dntps – 0.6 × n (no. of samples)
7. Taq polymerase – 1 × n (no. of samples)
Master mix for 95th primer:
1. Milli Q – 2.2 × n (no. of samples)
2. PCR Buffer – 1 × n (no. of samples)
3. MgCl2 – 0.8 × n (no. of samples)
4. Forward primer – 0.2 × n (no. of samples)
5. Reverse primer – 0.2 × n (no. of samples)
6. Dntps – 0.6 × n (no. of samples)
7. Taq polymerase – 1 × n (no. of samples)
Master mix for 82nd primer:
1. Milli Q – 2.2 × n (no. of samples)
2. PCR Buffer – 1 × n (no. of samples)
3. MgCl2 – 0.8 × n (no. of samples)
4. Forward primer – 0.2 × n (no. of samples)
5. Reverse primer – 0.2 × n (no. of samples)
6. Dntps – 0.6 × n (no. of samples)
7. Taq polymerase – 1 × n (no. of samples)
PCR conditions :-
Mt DNA primer:- [ M23, M15, M12 ]
• 95o C– 5 min.
• 95o C– 30 sec.
• 58o C– 30 sec.
• 72o C– 3 min.
• 72o C– 7 min.
• 4o C- ∞
• 35 cycles
Y DNA primer:- [ M95, M82]
•
96o C– 5 min.
•
94o C– 1 min.
•
54o C– 1 min.
•
72o C– 1 min.
•
72o C– 5 min.
•
4o C- ∞
•
35 cycles
Gel Electrophoresis of PCR products :-
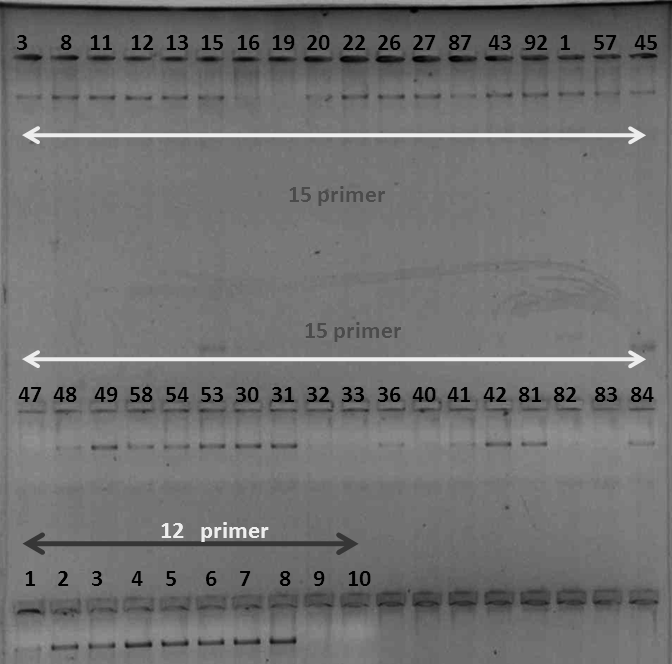
Gel check was done for each amplified PCR product.
Fig 20 : Gel Check of PCR products
These amplified products were stored at 4oC .
Sequencing of PCR products :-
1µl of PCR product was added to each well of sequencing plate.
1/3rd of the primer concentration [a] used for PCR × no. of samples was
calculated as [b].
Any one of the primer either forward or reverse was used.
Milli Q = ( 2.2 – [a] ) × no. of samples was added to [b].
The resulting mixture of primer and milli Q was added 2.2 µl to each well.
175 µl of Sequencing buffer + 25µl of Big Dye was prepared into a
mixture.
1.8 µl of the Dye Buffer mix was added to each well.
The plate was centrifuged at 12000 rpm for few seconds.
The plate was covered properly with alcohol washed flap and kept for
sequence PCR.
Conditions for sequence PCR:-
• 96o C– 10 sec.
• 55o C– 5 sec.
• 60o C– 4 min.
• 4o C- ∞
• 30 cycles
Processing :-
120µl of Sodium Acetate + 3 ml of absolute alcohol was made into a
mixture.
25µl of the above mixture was added to all the wells of sequencing plate.
It was incubated at room temperature for 10 min.
The plate was wrapped in tissue roll.
It was centrifuged at 4000 rpm at 16oC for 16 min.
The plate was gently inverted to discard sodium acetate mixture.
100 µl of 80% alcohol [ 8ml alcohol + 2 ml Milli Q]



















































