The Probation
If honours year is a sprint, then PhD is a marathon. There is no doubt about it. If anyone was to do PhD like in honours year, he will really collapse and burn out very fast.
I can never pinpoint the exact time I started my PhD other than the date I wrote on my application – 15th July 2004 – which came and gone without me noticing. Neither did I remember a point where anyone said to me in effect of “you start your PhD as of today.” However, there was a point in time where all these clicked in, and at the most unlikely place – immigration office at Casselden Place, at the end of Lonsdale Street. I had an immigration appointment in early July where I found out that I will need to re-do all the medical checkups for visa all over again and my next appointment will be after 15th July. I asked the immigration office if it will be alright if I were to start my PhD on 15th July as planned which was before I got my visa. He said “no problem, just go and start on your PhD first.” This very reply jotted me to realize that I am indeed starting my PhD.
Both Mary and Kevin are consistent about one thing about PhD – I am to start the candidature as a student and end it as a colleague, not an additional skilled pair of hands. I am to develop independent thoughts. If there is no quarrel, it is a sign that the supervision has failed. Kevin had drilled into me very early that at the end of the day, it is my thesis – I have to develop a thesis. The latin name for PhD is Philosophiae Doctor or Doctor of Philosophy (DPhil) in English – I have to develop my own philosophy. Only then, I will be worthy of the title of doctor.
At around that time, I had moved from Elsteinwick to Flemington / Newmarket, about 5 minutes walk from Laksa King, a nice Hong Kong / Malaysian eatery which was very popular with Southeast Asian students. I lived there for one and a half years until I came back to Singapore to do my internship. I never actually ate at Laksa King during my entire stay there. It was always “I can eat at Laksa King tomorrow” but that tomorrow never came.
I lived in Flemington for about one and a half years, with Michael Loke and his girlfriend (Juliet) as my housemate for the last year. Michael and myself crossed path due to our shared interest in artificial intelligence and that was prior to the inaugural AUSCC. I was in Sydney between the end of my honours year and the start of my PhD when Michael was asking me how to deal with the emotions of asking a girl to be his girlfriend. I remembered giving him both route of actions for acceptance and rejection – if he got accepted, all he needs to do is to jump around his block for a few rounds until he got exhausted. But if he got rejected, well, there will be someone else for him. The last thing I know about their relationship is that Juliet is now his wife.
During the entire stay in Flemington, I walked to school for most of the days – it took about 16 thousand steps one way and about 45 minutes. I found that the walk helps me to clear my mind and think.
In order to make some money to cover for the shortfall in living expenses, Mary recommended me to take up a practical for first year biology which I agreed. It was a practical on developmental biology – to show the development of Xenopus (African frog). Upon the approval of Graeme Campbell, then the Professor of Zoology, I became the Head Demonstrator for that practical. I remembered Graeme as a man of a giant statute with a long pony-tail but very friendly and kind. Based on reliable sources, he loved to talk about sex in class – isn’t that what biologists love to talk about? Probably the most amazing thing was what he did right after his retirement – have another child!!
There were 1300 biology undergraduates back then and I had to repeat the same practical for 15 times, twice a day, 3 hours per session, over 2 weeks. I can assure you that I really do not want to talk after that. In fact, I remembered walking back to office after the first week and met Mary on the corridor. I just told her off – “don’t talk to me.” She understood.
Having said that, it was a great experience and the money was great. I was paid an average of AUD 65 per hour. That was almost 3000 bucks for 2 weeks of work – my best paid job by hourly rate even up to today.
When I started my PhD, I did not know what will be my topic except that it is in bioinformatics. I ended up with a grandiose idea which is grandiose even today. I wanted a simulate-able mammary gland or cell and populate the simulation data and equations from published literature which can be used as a dry-lab equivalent of experimental platform for Kevin’s group.
In my plan to Dairy CRC, in which I illustrated my grandiose idea, there are 3 stages of my work (proposal in Appendix B):
-
MouseWay – a generic network of mouse biochemistry pathway extracted from mouse-specific literature. All pathways are homogeneous and there will be no compartmentalization or cellular location in MouseWay. This will be a simulate-able mouse cell.
-
Refinement of MouseWay to include compartmentalization, cellular location and enzymatic rates whenever possible.
-
MurioLactoLand – transformation of MouseWay for mammary-specificity by removing pathways and reactions that are non-existent in the mouse mammary gland and add missing mammary-specific pathways from the literature.
I presented this idea in the first lab meeting and Sonia commented that this will be a lifetime of work rather than a PhD. She is wrong – it will be a few lifetimes of work as I realized later – something that can be comparable to the holy grail of lactation biology. On hindsight, my final PhD thesis is really a subset of MouseWay.
Moving out of honours room but there was no space in postgraduate room, which is just opposite honours room. Hence, I was temporarily held in a lab space at 2nd floor of Zoology building with Brandon Menzies, Marilyn’s PhD student, then Nanette, another of Marilyn’s PhD student from Germany joined the small room. We got along pretty well. One afternoon, I did not know what happened but we were told to evacuate the room and to use the open lab space at 2nd floor. Well, we knew that the room was temporary and certainly not ideal – looked a little like a quasi-storage area – but at least it has a locked door and we can safely leave our laptops there. There was no security of any kind in the open lab area and we have to move out of the room by the end of the day. Brandon was not happy and pretty stressed. I was very distressed and fired a desperate email to Mary Familari, who was also a postgraduate coordinator, at about 3pm that day. Mary immediately flew into action and got Peter Krotsis, the department laboratory and safety manager, to look into the situation. By 4pm or so, all 3 of us secured a space in postgraduate room.
I attacked my project from 2 different ways – modelling and simulation, and extracting protein-protein interactions from published literture. I started on the modelling and simulation first. By then and due to my honours year, I had more experience with Python programming.
My difficulty then was the lack of knowledge in biological modeling and simulation, as well as text mining. However, I am determined to do something. I remembered that I had borrowed a textbook on natural language processing from the library (Speech and Language Processing by Daniel Jurafsky and James H. Martin) and vowed to fast until I finished the entire book of about 500 pages. I did that in 5 days and only started eating on the 6th. This is the basis I had for text mining.
I attacked the modeling and simulation aspect by looking at the currently available tools. Then, Systems Biology Markup Language (SBML) is gaining popularity while CellML is dying. I examined SBML and did not find it to be satisfactory for my use. My main reason is this. SBML model is based on functional categories. All the component definitions in a model are grouped together and the kinetic equations are grouped together. To me, this means that I am not able to abstract out different parts of the model easily. For example, I cannot easily remove a pathway from a complex cellular model unless I am willing to read the entire SBML code and remove the parts individually. SBML is function-oriented rather than object oriented. As such, my first plan was to develop an object-oriented modeling language and a simulator based on it. The result was a language called Mosirium Codes for Modeling and Simulation (MCMAS, see Appendix C) and “Mosirium” is my abbreviation for “mouse simularium”. I had even written a parser for MCMAS and a very rudimentary simulator for it. My intention was to publish MCMAS, then an interface between SBML and MCMAS, followed by a manuscript on Mosirium itself. However, ACM Transactions of Programming Languages and Systems rejected my manuscript on MCMAS on the basis that it did not solve any of the current problems in simulation. Looking back, I was pretty naïve then about the publishability of my work. It was partly due to the fact that I had not gotten a manuscript rejection then. It was around that time that I came across Kouchi Takahashi’s PhD project on ECell-3, which was done in Keio University, Japan, and started communicating with him. That was eventually how I got to visit Keio University.
By then, I had gotten a little headway into the text mining aspect and stopped working on MCMAS/Mosirium since. The only public relic of MCMAS and Mosirium was my poster in the 3rd Asia-Pacific Bioinformatics Conference. I had been considering about restarting this work for a long time but never got to it at all up to today and I do not foresee going anywhere with this unless there is a real need to. Although I still consider MCMAS to be a feasible idea, I realized that going against the scientific norm without substantial evidence is very tough and perhaps, not productive. At the same time, I also realized my severe lack of background knowledge in simulation and the required mathematics while reading Kouchi’s thesis. That really made me re-consider the feasibility of such approach but it was not until the later half of 2005 that I stopped toying with MCMAS and Mosirium after realizing that it was just too much for me to handle on my own. The concept of MCMAS and Mosirum was still in my confirmation report. I started to proceed on text mining with full steam.
One of the biggest lessons learnt from the MCMAS experience was not to attempt to re-invent the wheel – it is just untested and certainly not novel enough for publications, not to mention that I will be spending far too much time on it. I realized that I am a user of technology with some inclinations towards technology discovery rather than technology discovery with an inclination towards application. As a biologist-bioinformaticist, I should be adapting existing technologies and innovating them for my work rather than developing the technologies myself. If I were to go back to the start of my PhD, I should have just used ECell-3. It will certainly be more efficient though I am not convinced that I will make many inroads then, as I looked back. The development of primary technologies should be left to the computer scientists, mathematicians and the like.
When I started with text processing, there were 3 choices – NLTK (a Python natural language processing toolkit), MontyLingua (a Python natural language processing system) and GATE (a Java natural language processing toolkit). I chose to go with MontyLingua as it is a full system and had been published. MontyLingua was a project of Hugo Liu in MIT media labs and I find that to be a good basis for me to learn about natural language processing.
By then, I had collected all the abstracts of mouse and rat papers from PubMed. It is interesting to realize that humans accounted for almost half of the papers (about 7 million) while there were only about 800 thousand papers on mouse and 1.2 million papers on rat respectively. I will think that there should be more mouse papers instead of rat. My task then was to deduce MAP kinase pathway from the papers as a proof of concept that my text mining pipeline is working and the result was the first case study from my thesis.
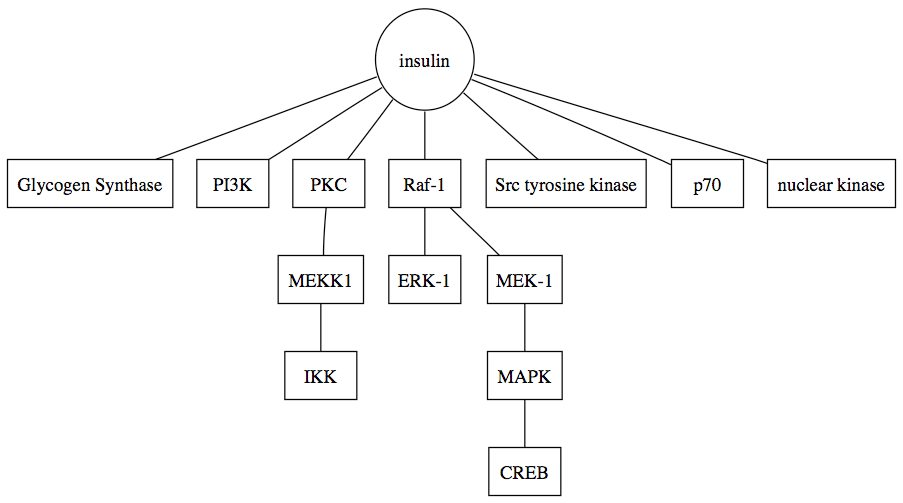
Extracted MAP kinase pathway (Source: Ling et al., 2007)
I started to call this project “Muscorian” which means “mouse librarian”. I had a chat with Kevin about the mouse versus rat issue and decided that I will consider them together instead of separate. This becomes a contentious point in my research which Terence (Terry) Fletcher, a senior lecturer in the department, pointed out as a matter of scientific curiosity after my confirmation seminar – how much of a rat is a mouse? In fact, is it possible to look from a helicopter view of what we know about mouse and rat collectively and see the differences? That may be an interesting research.
When I started to look into text analysis, I had also started to look at the expertise in text analysis in the university and came across Andrew Lonie, a lecturer in the department of information system, who was working on the kidney project. And for the formality of confirmation, I will need a person from another department to be in my confirmation committee and Andrew became that person.
For the entire first year of my PhD, I was really struggling with the idea of what determines a pass in a PhD. On one hand, I am confident that Kevin and Christophe are capable supervisors who will point me to the right direction but on the other hand, I was really worried that their area of expertise are not within what I am working on. Kevin is an excellent lactation and mammary biologist and Christophe’s work in mainly on sequence analysis and microarrays. I was terribly concerned. At the same time, my second upper honours results still hung over my head. The real question is whether am I good enough? What if my best is not good enough? Just like in my honours year – my best was just not good enough for me to get a first class honours. Will this happen again for my PhD? Will my best be still off the mark? I just do not know what is good enough for a PhD. As such, I fired a long email to Mary airing my concerns the day before my confirmation talk. David MacMillian was notified of my concerns. I might have came out too blantly and it seemed to be a concern of my supervision from Kevin and Christophe. David suggested that Andrew be one of my supervisors for me to get more support on the text mining side. I think that was the main reason for Andrew’s involvement. And with the confirmation papers signed off, I lose my probationary status and gained a confirmed status.
There were a number of other events in my first year. The most important is the award of Melbourne International Fee Remission Scholarship (MIFRS). With this inflow of cash, I decided on two things. Firstly, I will want to contribute back to the society and was finding a way to do so. Lauren, a girl who was doing her honours in Kevin’s lab at that time, introduced me to the idea of sponsoring a child. I thought that was a good idea and started to look up on WorldVision. Before long, I decided to sponsor a child as a way to give my gratitude to the society and had been sponsoring since then.
The second thought was to finish up my degree in computing since I was only left with 3 more modules and my honours project to get a full honours degree in computing with University of Portsmouth through distance learning. With that, I contacted Informatics Computer School again, the private education provider of my Advanced Diploma in Computing, to finish up my degree. There was also a financial reason to this. Back in 2000, I had dropped out of the degree to do an advanced diploma due to my National Service and a lot of unpleasantries back then. However, I had paid for a degree programme. Hence, I was really trying my luck to see if I can use my “unused cash” by topping up the additional cash for it. Well, it turned out that the school agreed to it and I was on the degree programme by mid-2005.




