5.1.2) Sequencing Approaches for NGS
5.1.2.1 ) Pyrosequencing
This method was published by Mostafa Ronaghi and Pal Nyren at the Royal Institute of Technology in Stockholm in 1996. It is non-electrophoretic, bioluminescence method which relies on the fact that on incorporation of a nucleotide on a growing nucleotide chain a pyrophosphate is released, which is then proportionally converted into visible light using a series of enzymatic reactions. As only one out of the 4 possible nucleotides A/T/G/C are added and available at any one time so that only one letter can be incorporated and the intensity of light determines if more than one letter is there in a row. The previously added nucleotide is degraded before the addition of second nucleotide, this process is repeated until the template DNA sequence is determined. Instead of using modified nucleotides this method manipulates DNA polymerase by limiting the dNTPs amount by single addition.
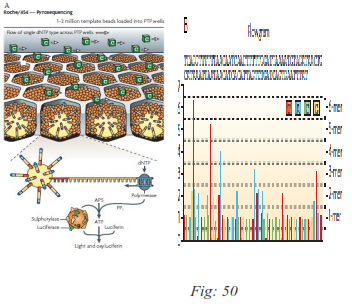
FIG: a Pyrosequencing using Roche/454’s Titanium platform. Following loading of the DNA-amplified beads into individual Pico Titer Plate (PTP) wells, additional beads, coupled with sulphurylase and luciferase, are added. In this example, a single type of 2′-deoxyribonucleoside triphosphate (dNTP) - cytosine - is shown flowing across the PTP wells. The fibre-optic slide is mounted in a flow chamber, enabling the delivery of sequencing reagents to the bead-packed wells. The underneath of the fibre-optic slide is directly attached to a high-resolution charge-coupled device (CCD) camera, which allows detection of the light generated from each PTP well undergoing the pyrosequencing reaction. B. The light generated by the enzymatic cascade is recorded as a series of peaks called a flowgram. PPi, inorganic pyrophosphate.
5.1.2.2 Sequencing by reversible terminator chemistry
This approach is based on reversible terminator which is bound to dNTPs, it is a cyclic method that comprises nucleotide incorporation, fluorescence imaging and cleavage of terminator. A fluorescently-labeled terminator is imaged as each dNTP is added and then cleaved to allow incorporation of the next base. These nucleotides are chemically blocked such that each incorporation is a unique event. An imaging step follows each base incorporation step, then the blocked group is chemically removed to prepare each strand for the next incorporation by DNA polymerase. This series of steps continues for a specific number of cycles, as determined by user-defined instrument settings. The 3' blocking groups were originally conceived as either enzymatic or chemical reversal.
5.1.2.3 Sequencing-by-ligation mediated by ligase enzymes:-
DNA ligase is an enzyme that joins together ends of DNA molecules. Sequencing by ligation relies upon the sensitivity of DNA ligase for base-pairing mismatches. The target molecule to be sequenced is a single strand of unknown DNA sequence. DNA ligase is sensitive to the structure of DNA and has very low efficiency when there are mismatches between the bases of the two strands.A short anchor strand is first brought in to bind the known sequence A mixed pool of probe oligonucleotides is then brought in (eight or nine bases long), labeled (typically with fluorescent dyes) according to the position that will be sequenced. These molecules hybridize to the target DNA sequence, next to the anchor sequence, and DNA ligase preferentially joins the molecule to the anchor when its bases match the unknown DNA sequence. Based on the fluorescence produced by the molecule, one can infer the identity of the nucleotide at this position in the unknownx.
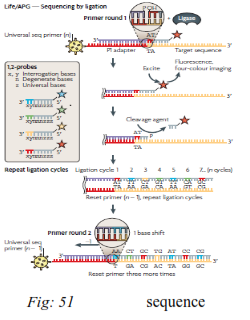
Sequencing by ligation can proceed in either direction (either 5'-3' or 3'-5') depending on which end of the probe oligonucleotides are blocked by the label. The 3'-5' direction is more efficient for doing
ribonucleases and acid, respectively. multiple cycles of ligation. Note that this is the opposite direction to polymerase based sequencing methods.
5.1.2.4) Phospholinked Fluorescent Nucleotides or Real-time sequencing
Single molecule real time sequencing (also known as SMRT) is a single molecule DNA sequencing by synthesis technology developed by Pacific Biosciences. Single molecule real time sequencing utilizes the zero-mode waveguide (ZMW). The ZMW is a structure that creates an illuminated observation volume that is small enough to observe only a single nucleotide of DNA (also known as a base) being incorporated by DNA polymerase. A single DNA polymerase enzyme is affixed at the bottom of a ZMW with a single molecule of DNA as a template. Each of the four DNA bases is attached to one of four different fluorescent dyes. When a nucleotide is incorporated by the DNA polymerase, the fluorescent tag is cleaved off and diffuses out of the observation area of the ZMW where its fluorescence is no longer observable. For each of the nucleotide bases, there are four corresponding fluorescent dye molecules that enable the detector to identify the base being incorporated by the DNA polymerase as it performs the DNA synthesis. The fluorescent dye molecule is attached to the phosphate chain of the nucleotide. When the nucleotide is incorporated by the DNA polymerase, the fluorescent dye is cleaved off with the phosphate chain as a part of a natural DNA synthesis process during which a phosphodiester bond is created to elongate the DNA chain. The cleaved fluorescent dye molecule then diffuses out of the detection volume so that the fluorescent signal is no longer detected. A detector detects the fluorescent signal of the nucleotide incorporation, and the base call is made according to the corresponding fluorescence of the dye.
5.1.3) Next generation DNA sequencers:
A DNA sequencer is a scientific instrument used to automate the DNA sequencing process. Given a sample of DNA, a DNA sequencer is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The order of the DNA bases is reported as a text string, called a read. Some DNA sequencers can be also considered optical instruments as they analyze light signals originating from fluorochromes attached to nucleotides.
5.1.3.1) Roche 454 Genome Sequencer FLX
Roche 454 was the first commercially successful next generation system. This sequencer uses pyrosequencing technology. In this system, the whole genome can be sequenced without any cloning step, Instead of using dideoxynucleotides to terminate the chain amplification, pyrosequencing technology relies on the detection of pyrophosphate released during nucleotide incorporation. The library DNAs with 454-specific adaptors are denatured into single strand and captured by amplification beads followed by emulsion PCR . Then on a picotiter plate, one of dNTP (dATP, dGTP, dCTP, dTTP) will complement to the bases of the template strand with the help of ATP sulfurylase, luciferase, luciferin, DNA polymerase, and adenosine 5 phosphosulfate (APS) andrelease pyrophosphate (PPi) which equals the amount of incorporated nucleotide. The ATP transformed from PPi drives the luciferin into oxyluciferin and generates visible light. At the same time, the unmatched bases are degraded by apyrase. Then another dNTP is added into the reaction system and the pyrosequencing reaction is repeated.
The most outstanding advantage of Roche is its speed: it takes only 10 hours from sequencing start till completion. ). One of the shortcomings is that it has relatively high error rate in terms of poly-bases longer than 6 bp
5.1.3.2) Illumina genome analyzer
It is based on the concept of Sequencing By Synthesis (SBS) technology also termed as Real-time sequencing. As explained earlier it starts with DNA fragments ligated with specific adapters and, after denaturation, the sequencing templates are immobilized at one end on surface of a glass flowcell which is coated thickly with the adapters and the complementary adapters. Each flowcell is divided into eight separate lanes, and the interior surfaces have covalently attached oligomers complementary to the specific adapters that are ligated onto the library fragments. Hybridization of these DNAs to the oligomers on the flow cell occurs by an active heating and cooling step, followed by a subsequent incubation with reactants and an isothermal polymerase that amplifies the fragments in a discrete area or ‘cluster’ on the flow cell surfaces. The flow cell is placed into a fluidics cassette within the sequencer, where each cluster is supplied with polymerase and four differentially labeled fluorescent nucleotides that have their 30-OH chemically inactivated to ensure that only a single base is incorporated per cycle. Each base incorporation cycle is followed by an imaging step to identify the incorporated nucleotide at each cluster and by a chemical step that removes the fluorescent group and deblocks the 30 end for the next base incorporation cycle. At the end of the sequencing run, the sequence of each cluster is computed and subjected to quality filtering to eliminate low-quality reads of between 32 and 40 bp.
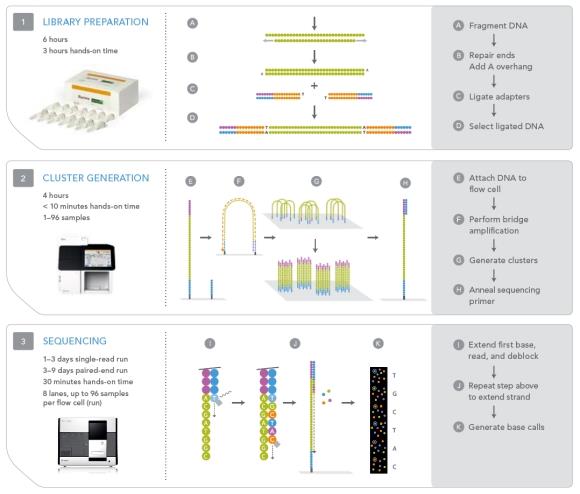
Fig: 51
Fig: Principle of Illumina genome analyzer
5.1.3.3). Applied Biosystems Solid sequencer
This instrument uses sequencing process catalyzed by DNA ligase, and employs emulsion PCR to amplifie each bead–DNA complex. After amplification, the beads are covalently attached to the surface of a specially treated glass slide that is placed into a fluidics cassette within sequencer. In the Solid system, two slides are processed per run; one slide receives sequencing reactants as the second slide is being imaged. The ligation-based sequencing process starts with the annealing of a universal sequencing primer that is complementary to the Solid specific adapters on the library fragments. The addition of a limited set of semi-degenerate 8mer oligonucleotides and DNA ligase is automated by the instrument. After the ligation step, a fluorescent readout identifies the fixed base of the 8mer, which corresponds to either the fifth position or the second position, depending on the cycle number. A subsequent chemical cleavage step removes the sixth through eighth base of the ligated 8mer by attacking the linkage between bases 5 and 6, thereby removing the fluorescent group and enabling a subsequent round of ligation. The process occurs in steps that identify the sequence of each fragment at five nucleotide intervals, and the synthesized fragments that end at base 25 (or 35 if more cycles are performed) are removed by denaturation and washed away. A second round of sequencing initiates with the hybridization of an n-1 positioned universal primer, and subsequent rounds of ligation-mediated sequencing, and so on.
Each SOLiD (Sequencing by Oligo Ligation and Detection) run requires 5 days and produces 3–4 Gb of sequence data with an average read length of 25–35 bp.
5.1.4) NGS application:
NGS is applied for the finding of non-virulence genes in bacterial and viral species. NGS has the ability to visualize RNA expression in sequence form hence it has replace microarray analysis.
The cost of diagnosis is reduced with NGS and a substantial increase in throughput and accuracy.







