How do cells divide?
There are two types of cell division: mitosis and meiosis. Most of the time when people refer to "cell division," they mean mitosis, the process of making new body cells. Meiosis is the type of cell division that creates egg and sperm cells.
Mitosis is a fundamental process for life. During mitosis, a cell duplicates all of its contents, including its chromosomes, and splits to form two identical daughter cells. Because this process is so critical, the steps of mitosis are carefully controlled by a number of genes. When mitosis is not regulated correctly, health problems such as cancer can result.
The other type of cell division, meiosis, ensures that humans have the same number of chromosomes in each generation. It is a two-step process that reduces the chromosome number by half-from 46 to 23-to form sperm and egg cells. When the sperm and egg cells unite at conception, each contributes 23 chromosomes so the resulting embryo will have the usual 46. Meiosis also allows genetic variation through a process of DNA shuffling while the cells are dividing.
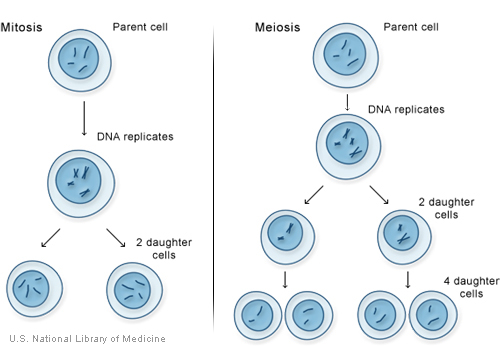
Mitosis and meiosis, the two types of cell division.
For more information about cell division:
For a detailed summary of mitosis and meiosis, please refer to the chapter titled What Is A Cell? (http://www.ncbi.nlm.nih.gov/About/primer/genetics_cell.html) In the NCBI Science Primer. Scroll down to the heading "Making New Cells and Cell Types."
How do genes control the growth and division of cells?
A variety of genes are involved in the control of cell growth and division. The cell cycle is the cell's way of replicating itself in an organized, step-by-step fashion. Tight regulation of this process ensures that a dividing cell's DNA is copied properly, any errors in the DNA are repaired, and each daughter cell receives a full set of chromosomes. The cycle has checkpoints (also called restriction points), which allow certain genes to check for mistakes and halt the cycle for repairs if something goes wrong.
If a cell has an error in its DNA that cannot be repaired, it may undergo programmed cell death (apoptosis) (illustration on page 29). Apoptosis is a common process throughout life that helps the body get rid of cells it doesn't need. Cells that undergo apoptosis break apart and are recycled by a type of white blood cell called a macrophage (illustration on page 29). Apoptosis protects the body by removing genetically damaged cells that could lead to cancer, and it plays an important role in the development of the embryo and the maintenance of adult tissues.
Cancer results from a disruption of the normal regulation of the cell cycle. When the cycle proceeds without control, cells can divide without order and accumulate genetic defects that can lead to a cancerous tumor (illustration on page 30).
For more information about cell growth and division:
The National Institutes of Health's Apoptosis Interest Group (http://www.nih.gov/ sigs/aig/Aboutapo.html) provides an introduction to programmed cell death.
The National Cancer Institute offers several publications that explain the growth of cancerous tumors. These include What You Need To Know About Cancer-An Overview (http://www.cancer.gov/cancertopics/wyntk/cancer) and Understanding Cancer (http://www.cancer.gov/cancertopics/understandingcancer/cancer).
Illustrations
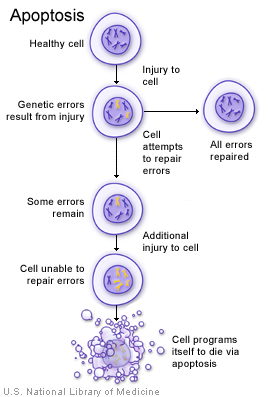
A damaged cell may undergo apoptosis if it is unable to repair genetic errors.
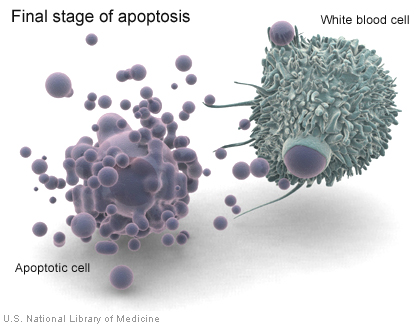
When a cell undergoes apoptosis, white blood cells called macrophages consume cell debris.
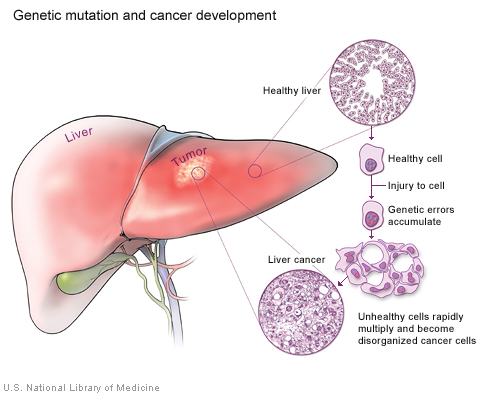
Cancer results when cells accumulate genetic errors and multiply without control.
How do geneticists indicate the location of a gene?
Geneticists use maps to describe the location of a particular gene on a chromosome. One type of map uses the cytogenetic location to describe a gene's position. The cytogenetic location is based on a distinctive pattern of bands created when chromosomes are stained with certain chemicals. Another type of map uses the molecular location, a precise description of a gene's position on a chromosome. The molecular location is based on the sequence of DNA building blocks (base pairs) that make up the chromosome.
Cytogenetic location
Geneticists use a standardized way of describing a gene's cytogenetic location. In most cases, the location describes the position of a particular band on a stained chromosome:
17q12
It can also be written as a range of bands, if less is known about the exact location:
17q12-q21
The combination of numbers and letters provide a gene's "address" on a chromosome. This address is made up of several parts:
• The chromosome on which the gene can be found. The first number or letter used to describe a gene's location represents the chromosome. Chromosomes 1 through 22 (the autosomes) are designated by their chromosome number. The sex chromosomes are designated by X or Y.
• The arm of the chromosome. Each chromosome is divided into two sections (arms) based on the location of a narrowing (constriction) called the centromere. By convention, the shorter arm is called p, and the longer arm is called q. The chromosome arm is the second part of the gene's address. For example, 5q is the long arm of chromosome 5, and Xp is the short arm of the X chromosome.
• The position of the gene on the p or q arm. The position of a gene is based on a distinctive pattern of light and dark bands that appear when the chromosome is stained in a certain way. The position is usually designated by two digits (representing a region and a band), which are sometimes followed by a decimal point and one or more additional digits (representing sub-bands within a light or dark area). The number indicating the gene position increases with distance from the centromere. For example: 14q21 represents position 21 on the long arm of chromosome 14. 14q21 is closer to the centromere than 14q22.
Sometimes, the abbreviations "cen" or "ter" are also used to describe a gene's cytogenetic location. "Cen" indicates that the gene is very close to the centromere. For example, 16pcen refers to the short arm of chromosome 16 near the centromere. "Ter" stands for terminus, which indicates that the gene is very close to the end of the p or q arm. For example, 14qter refers to the tip of the long arm of chromosome
14. ("Tel" is also sometimes used to describe a gene's location. "Tel" stands for telomeres, which are at the ends of each chromosome. The abbreviations "tel" and "ter" refer to the same location.)
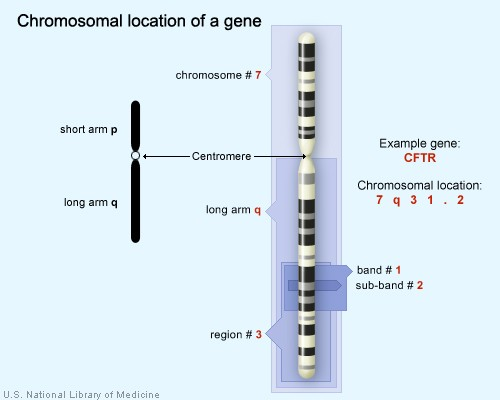
The CFTR gene is located on the long arm of chromosome 7 at position 7q31.2.
Molecular location
The Human Genome Project, an international research effort completed in 2003, determined the sequence of base pairs for each human chromosome. This sequence information allows researchers to provide a more specific address than the cytogenetic location for many genes. A gene's molecular address pinpoints the location of that gene in terms of base pairs. It describes the gene's precise position on a chromosome and indicates the size of the gene. Knowing the molecular location also allows researchers to determine exactly how far a gene is from other genes on the same chromosome.
Different groups of researchers often present slightly different values for a gene's molecular location. Researchers interpret the sequence of the human genome using a variety of methods, which can result in small differences in a gene's molecular address. Genetics Home Reference presents data from NCBI (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene) for the molecular location of genes.
For more information on genetic mapping:
The National Human Genome Research Institute explains how researchers create a genetic map (http://www.genome.gov/10000715).
The University of Washington provides a Cytogenetics Gallery (http://www.pathology.washington.edu/galleries/Cytogallery/main.php?file=intro) that includes a description of chromosome banding patterns (http://www.pathology.washington.edu/galleries/Cytogallery/main.php?file=banding+ patterns).
The NCBI Science Primer offers additional detailed information about genome mapping (http://www.ncbi.nlm.nih.gov/About/primer/mapping.html). NCBI also provides information about assembling and annotating the genome (http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=handbook&part=ch14).








