5.3 ) Fluorescent in-situ Hybridization
Fluorescence in situ hybridization (FISH) is a powerful technique used in the detection of chromosomal abnormalities. FISH is a highly sensitive and specific Assay with a very high speed so it emerges as a pivotal cytogenetic technique that has provided significant advances in both the research and diagnosis of hematological malignancies and solid tumors.
Fluorescence in situ hybridization (FISH) is widely used for the localization of genes and specific genomic regions on target chromosomes, both in metaphase and interphase cells. FISH has gained importance in clinical practice because of the availability of relevant information that is otherwise difficult or impossible to obtain.
The technique employs fluorescent probes for the detection of specific nucleotide sequences in chromosomes. FISH has a much higher rate of sensitivity and specificity than other genetic diagnostic tests such as karyotyping and thus can be used to detect a variety of structural abnormalities in chromosomes, including small genetic deletions involving just one to five genes.
FISH also provides results more quickly than karyotyping because no cell culture is required. It is commonly used for preimplantation genetic diagnosis (PGD) during in vitro fertilization. PGD involves obtaining a single cell from an embryo in the blastocyst stage of development. This single cell can then be analyzed using FISH.
5.3.1) Principle of FISH
The success of FISH, and all other methods of in situ hybridization, depends on the remarkable stability of the DNA double helix. As in 1953 James Watson and Francis Crick revealed the story of double helical structure of DNA which makes detection technique possible to diagnose several chromosomal abnormalities.
The hydrogen bonds can be broken when treated with chemicals or heat which leads to disruption of helix structure but when the conditions become favorable the helix revert to reform. This ability of the DNA helix to re-form or renature, provides the basis for molecular hybridization with Probe.
The probes are complementary to specific parts of a chromosome. When DNA is heated, the patient’s two DNA strands break apart, or denature, and the probes are able to hybridize to their complementary sequence in the patient’s DNA.
In the 1960s, researchers Joseph Gall and Mary Lou Pardue realized that molecular hybridization of probes could be used to identify the position of DNA sequences in situ, they ublished a landmark paper demonstrating that radioactive copies of a ribosomal DNA sequence could be used to detect complementary DNA sequences.
The earliest in situ hybridizations, performed in the late 1960s, were not fluorescent at all, but rather utilized probes labeled with radioisotopes. Techniques not employing fluorescence, such as enzyme-based chromogenic reporters (reviewed by Hougaard et al., 1997) and gold-based probe systems used in electron microscopy (reviewed by Puvion-Dutilleul and Puvion, 1996) are each fields in their own right.
5.3.2) Procedure of FISH and DNA PROBE preparation:
With FISH different types of sequences of the human genome are detectable. The euchromatic region of an entire chromosome or chromosome arm can be visualized by whole chromosome paints (wcp).
Probes:
The genome contains a mixture of DNA sequences that is repetitive at some points and shows some unique sequences. Some of the repetitive sequences are located in compact clusters at particular locations in the genome such as centromeres, while others are interspersed among unique sequences . The centromeric regions contain stretches of randomly repeated short sequences, of many mega-bases. These belong to several repetitive sequence families, groupings that share high-sequence similarity. There is sufficient difference in portions of this quincesa family so that probes that hybridize with predominant specificity to one chromosome centromeric region have been obtained for almost all human chromosomes.
Probes can be produced that hybridize to all centromeric regions. The presence of these interspread repeated consequetive sequence makes it possible to use FISH to stain Genomic sequence. Gene probes are generally longer than 500 bases and comprise all or most of a target gene.They can be generated in two ways. Cloned probes are normally used when a specific clone is available or when the DNA sequence is unknown and must be cloned first in order to be mapped and sequenced. It is usual to cut the gene with restriction enzymes and excise it from an agarose gel, although if the vector has no homology, this might not be necessary.
The majority of radioactive labeling procedures rely upon enzymatic incorporation of a nucleotide labeled into the DNA, RNA, or oligonucleotide.
FISH probes are generally made out of BAC clones with genomic DNA containing a variable amount of repetitive DNA that will need to be removed or blocked for FISH analysis . To generate repeat free (RF) Probes without loss in genomic coverage, a random library is made from BAC clones by whole-genome amplification (WGA
5.3.3) Labeling and Detection probes
Fluorophores can be divided into three general groups:
- Organic dyes
- Biological fluorophores
- Quantum dots
Each fluorophore has distinct characteristics, which should be considered when deciding which fluorophore to use for a given application or experimental system. Organic dyes
Synthetic organic dyes, such as fluorescein, were the first fluorescent compounds used in biological research. Derivatives of these original compounds have been produced to improve their photostability and solubility. These dyes are also derivitized to use in bioconjugation, especially fluorescein isothiocyanate (FITC), rhodamine (tetramethylrhodamineisothiocyanate, TRITC) and commercial variants with greater performance.
Biological fluorochrome
While bioluminescence has been known for millenia, the first use of a biological fluorophore for research applications occurred in the 1990s, when green fluorescent protein (GFP) was cloned from the jellyfish Aequoreavictoria and used as a gene expression reporter (3). Since that time, derivatives of the original GFP, phycobiliproteins (allophycocyanin, phycocyanin, phycoerythrin and phycoerythrocyanin) and many other proteins have been designed for use in biological expression systems, and their use is now commonplace in biological research.
Quantum dots
Quantum dots are nanocrystals with unique chemical properties that provide tight control over the spectral characteristics of the fluor. Quantum dots were developed in the 1980s and since the 1990s have been increasingly used in fluorescence applications in biological research. Quantum dots are Nano scale-sized (2-50nm) semiconductors that, when excited, emit fluorescence at a wavelength based on the size of the particle; smaller quantum dots emit higher energy than large quantum dots, and therefore the emitted light shifts from blue to red as the size of the nanocrystal increases. And because quantum dot size can be tightly controlled, there is greater specificity for distinct excitation and emission wavelengths than other flurochromes.
5.3.4 ) Types of Label
I) Radioactive Labels:
Nucleic acid probes can be labeled using radioactive isotopes. Radiolabeled probes used to be the most common type but are less popular today because of safety considerations as well as cost and disposal of radioactive waste products. However, radiolabeled probes are the most sensitive, as they provide the highest degree of resolution currently available in hybridization assays.
II) Nonradioactive Labels:
Compared to radioactive labels, the use of nonradioactive labels have several advantages:
- Safety.
- Higher stability of probe.
- Efficiency of the labeling reaction.
- Detection in situ.
- Less time taken to detect the signal.
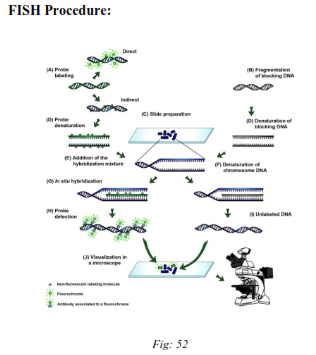
Fig: Fluorescent In-situ Hybridization technique.
With FISH different types of sequences of the human genome are detectable. The euchromatic region of an entire chromosome or chromosome arm can be visualized by whole chromosome paints (wcp).
These probes contain clones of DNA libraries of the specific chromosome. Centromere specific probes consist of highly repetitive sequences (a satellite DNA) that are present in the centromere of each chromosome.
Procedure of fluorescence in situ hybridization (FISH) consists of four fundamental steps:
- Denaturation of chromosome
- Hybridization of probe-DNA to the existing single strands DNA to form complementary double strands (renaturation) in several hours (4-16 h) at 37° C.
- Detection of the hybridized sequences is required, according to the fact that the DNA probe already contains nucleotides linked to a fluorescent dye (direct labeling) or if so-called immunogenic reporter molecules such as biotin or digoxigenin are applied, being accessible to specific antibodies that are conjugated to a fluorochrome (indirect labeling).
The fluorescent dyes most frequently applied in both methods are fluorescein isothiocyanate (FITC, yellow) and tetra-methyl rhodamin isothiocyanate (TRITC, red). By using two different reporter molecules labeled with different fluorescent dyes several regions in the genome can be analyzed simultaneously.
5.3.5 ) Application of Interphase FISH in Clinical Practice
One of the most important prerequisites for prenatal diagnostics is minimal risk as well as quick, valid and reliable results. The advantage of interphase FISH in this context is the rapid detection of selected chromosomal abnormalities, typically the ploidy status of chromosomes
Postnatal Diagnostics
Interphase FISH is suitable for postnatal analyses on all tissues containing nucleated cells that can be prepared. No cultivation is required, so fixed tissues can be analyzed as well as material that is difficult to cultivate
Tumor Diagnostics
The number of mitoses is low according to the type of tumor and chromosomes frequently are displayed in such a resolution that will not enable unequivocal structural analysis.
Interphase hybridization, therefore, turned into an important component in diagnosis as well as prognosis and therapy of leukemia, lymphoma and solid tumors. Essentially, tumor characteristic chromosome disorders (aneuploidies, translocations, deletions) can be analyzed as well as the amplification of specific genes.
Fluorescence in situ hybridization (FISH) is widely used to describe bacterial community composition and, to a lesser extent, to describe the physiological state of cells. One of the limitations of the technique is that the effectiveness of the detection of target cells appears to vary widely. FISH not only allows the detection of culturable microorganisms, but also of yet-to-be cultured (so-called unculturable) organisms, and can therefore help in understanding complex microbial communities.




