5.4 ) QF-PCR
Over the last few decades, for prenatal diagnosis of fetal chromosomal abnormalities prenatal cytogenetic analysis of cultured amniocytes, chorionic villi, or fetal blood is done. Aneuploidy, or the loss or gain of a chromosome in a diploid complement is a common cause of human genetic disorders. Pregnancies with chromosomal aneuploidies that survive to term including trisomies of autosomal chromosomes 13, 18 and 21 constitute approximately 90 percent of chromosomal abnormalities with a severe phenotype. Trisomies can thus be divided into four categories based on the size of the triplicated genomic region. They are complete (whole-chromosome) trisomies resulting from meiotic or mitotic non-disjunction that account for up to 0.5% of live births; partial (segmental) trisomies involving a genomic region of more than one chromosomal band (usually larger than 5 Mb) resulting from abnormal meiosis and segregation in individuals with balanced chromosomal rearrangements which is much less frequent than whole-chromosome trisomies; microtrisomies (segmental duplication) that are partial trisomy of a genomic segment that is shorter than 3–5 Mb; and the duplications of only one gene or one functional genomics element that can also be pathogenic.
Trisomy 18, also known as Edward syndrome, first described in 1960, is the most common trisomy behind Down syndrome, occurring in 1:3000 conceptions and 1:6-8000 live births.
Quantitative fluorescent polymerase chain reaction (QFPCR) has recently entered the field of prenatal diagnosis to overcome the need to culture fetal cells, hence to allow rapid diagnosis of some selected chromosomal anomalies. We reviewed the studies on the accuracy of QF-PCR in detecting chromosomal anomalies at prenatal diagnosis. Overall, 22 504 samples have been analysed. QF-PCR might play a major role and be considered a valid alternative to the full karyotype. Being less expensive, and almost entirely automated, more women could undergo invasive prenatal diagnosis without significant increase in health expenditure.
This disorder shows a higher incidence for females believed to be due to a higher prenatal mortality of males. Trisomy 13, also called Patau syndrome, though of rare occurrence with an incidence of 1 in 16,000 live births, causes much more severe and life threatening medical conditions that allows only about 5 to 10 percent of the affected children to live up to the first year of their life.
This will have the benefit of providing rapid and accurate results to women at increased risk of fetal Down syndrome, trisomy 13, trisomy 18, sex chromosome aneuploidy or triploidy. It will also promote better use of laboratory resources and reduce the cost of prenatal diagnosis. QF-PCR is a reliable method to detect trisomies and should replace conventional cytogenetic analysis whenever prenatal testing is performed solely because of an increased risk of aneuploidy in chromosomes 13, 18, 21, X or Y. As with all tests, pretest counselling should include a discussion of the benefits and limitations of the test. In the initial period of use, education for health care providers will be required
5.4.1 ) Principle:
QF PCR analysis includes amplification, detection and analysis of chromosome-specific DNA sequences known as genetic markers or small tandem repeats (STRs). Fluorescently labeled marker specific primers are used for PCR amplification of individual markers and the copy number of each marker is indicative of the copy number of the chromosome. The resulting PCR products may be analyzed and quantified using an automated genetic analyzer.
The genetic markers/STRs may vary in length between individual chromosomes and subjects, depending on the number of repeated STRs. The relative copy number of each allele is determined by calculating the ratio of the peak areas or peak heights detected for each marker.
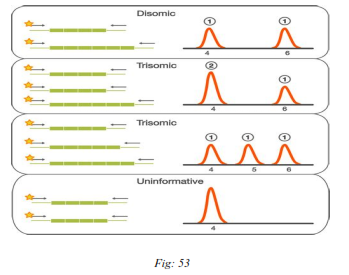
A normal diploid sample has the contribution of two of each of the investigated chromosomes. Two alleles of a chromosome specific marker are detected as two peaks in a 1:1 ratio when the marker is heterozygous and as one peak when the marker is homozygous (have alleles of same length). The detection of an additional allele as three peaks in a 1:1:1 ratio or as two peaks in a 2:1/1:2 ratio indicates the presence of an additional marker copy possibly corresponding to an additional chromosome, as in the case of trisomy. Subjects who are homozygous or monosomic for a specific marker will display only one peak.
Although currently QF-PCR would be considered the technology of choice for rapid aneuploidy detection, it is likely that other platforms will be developed that may have the capacity to detect additional abnormalities such as cases of segmental aneuploidies. For example, a bead array approach with bacterial artificial chromosomes— containing microbeads with probes for the aneuploidies and microdeletion syndromes has been developed and validated on a small number of samples.
The clinical application of quantitative fluorescent polymerase chain reaction (QF-PCR) for rapid prenatal detection of chromosome aneuploidies has been limited in most studies to the detection of autosomal trisomies. The assay proved to be so efficient and reliable that in most aneuploidy cases, in which ultrasound findings were in agreement with the molecular result, therapeutical interventions were possible without waiting for the result of cytogenetic analysis.
Germline aneuploidy is typically detected through karyotyping, a process in which a sample of cells is fixed and stained to create the typical light and dark chromosomal banding pattern and a picture of the chromosomes is analyzed. Other techniques include Fluorescence In Situ Hybridization (FISH), quantitative PCR of Short Tandem Repeats, quantitative fluorescence PCR (QF-PCR), quantitative PCR dosage analysis, Quantitative Mass Spectrometry of Single Nucleotide Polymorphisms, and Comparative Genomic Hybridization (CGH).
These tests can also be performed prenatally to detect aneuploidy in a pregnancy, through either amniocentesis or chorionic villus sampling. Pregnant women of 35 years or older are offered prenatal diagnosis because the chance of chromosomal aneuploidy increases as the mother's age increases.
Multiplex quantitative fluorescence PCR (QF-PCR) provides the possibility to detect copy number variation of chromosomal sequences in several hours. It also has the advantage of being much cheaper and allowing the simultaneous processing of larger numbers of samples than FISH and karyotyping analysis. However, the presence of multiple primer pairs in a PCR reaction reduces the reliability of the quantification.
To solve these technical problems, multiplex ligation-dependent probe amplification (MLPA) has emerged as an alternative to standard PCR-based techniques for detection of the chromosome aneuploidies




