Chapter One
1. BITTER–SWEET STORY OF THE IGF
RECEPTORS IN CELL (MAL)FUNCTIONING
Dragana Robajac{1}, Miloš Šunderić, Nikola Gligorijević, Olgica Nedić
Institute for the Application of Nuclear Energy (INEP),
University of Belgrade, Belgrade, Serbia
ABSTRACT
The insulin–like growth factor (IGF) system contains multiple members including growth factors, their binding proteins and receptors. After binding of growth factors to their receptors, a cascade of signals is activated initiating a number of mitogenic and metabolic pathways. Being at the crossroad of different and sometimes opposing functions, dependent on structural modifications as well as cell surrounding, the IGF system represents an intriguing field of investigation. It is involved in cell growth, proliferation and energy metabolism, but also in cell apoptosis. The IGF system will be described in the following chapter, with the focus on the IGF receptors and functions associated with them. The data on membrane proteins, their N– glycome and oxidation status will be related to our findings on the receptors in different physiological and pathological conditions, such as normal and abnormal tissue growth and development. Placental and colorectal tissues will be used as examples.
Keywords: insulin–like growth factors, membrane receptors, placenta, diabetes, colon cancer
1.1. INTRODUCTION
The insulin–like growth factor (IGF) system consists of peptides (IGF–I and IGF–II), binding proteins (IGFBP–1–6), receptors (IGF–1R and IGF–2R) and IGFBP proteases, as presented in Figure 1. Being closely related to IGF peptides and IGF–1R, and due to a high degree of homology and crossreactivity, insulin and insulin receptors (IR) can also be observed as the part of the IGF system, as will be discussed in the following sections.
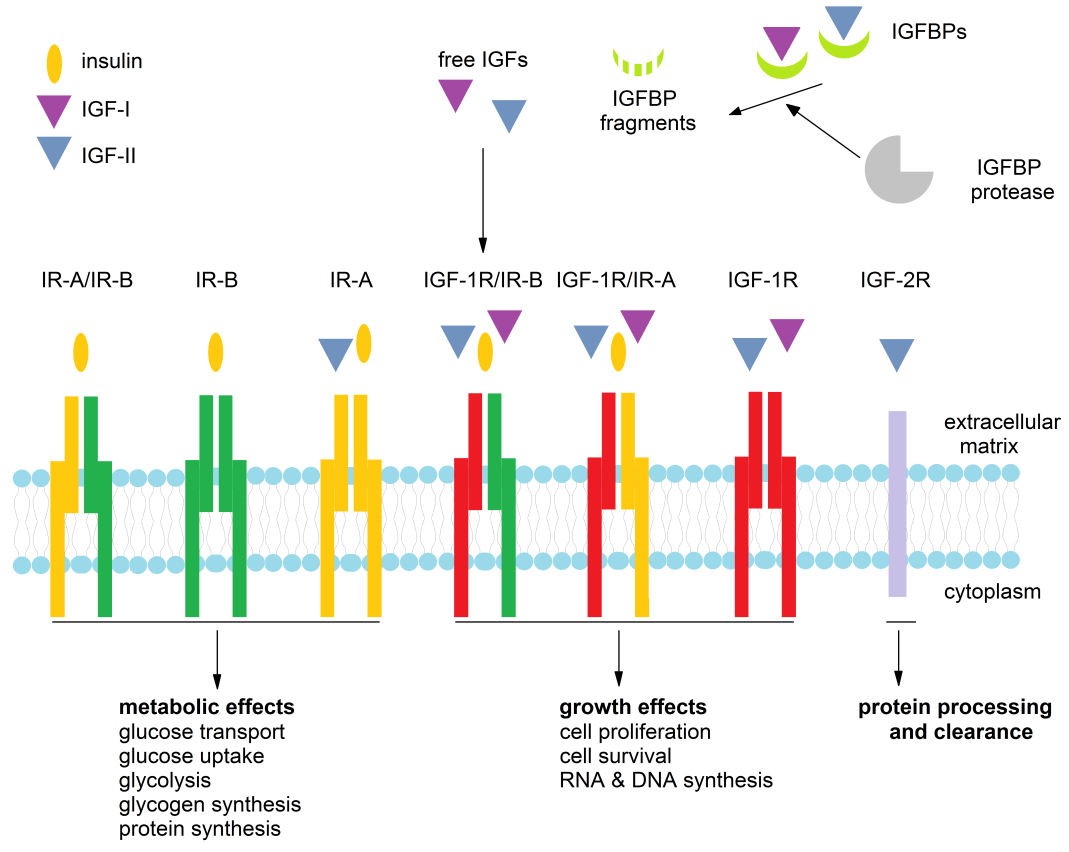
Figure 1. Members of the IGF system and the processes they control.
1.1.1. Peptides of the IGF system
Peptides of the IGF system are 7.5 kDa proteins, with amino acids arranged in four domains (B, C, A and D), and are mainly produced in the liver (Le Roith et al., 2001). IGF–I and IGF–II share 70% similarity, while the difference between IGFs and insulin is around 50% (Rinderknecht and Humbel, 1978). The main difference between IGFs and (pro)insulin is the existence of domains C and D (both are omitted from insulin while proinsulin contains domain C). A second difference originates from the specific amino acids in the IGFs located at positions 3, 4, 15 and 16, that are responsible for recognition and binding to IGFBPs. A third difference between IGFs and insulin is derived from the existence of IGFBPs, which bind IGFs leaving rather minute amounts of free peptides, whereas binding proteins do not control insulin availability, allowing this peptide to be able to circulate freely (Annunziata, Granata and Ghigo, 2011).
The IGF–I gene is located in the 12q region, and after birth its expression (and the production of IGF–I responsible for growth and proliferation) is under the control of growth hormone. The IGF–II gene is located in the 11p region, whose anomalies result in abnormal foetal and postnatal growth, as well as increased risk of embryonic tumours in early life (Wakeling et al., 2017; Brioude et al., 2018). The term genomic imprinting is used to describe the monoallelic expression of genes based on their parental origin (Cassidy and Charalambous, 2018). Genomic imprinting is found to be of high importance as imprinting disorders share one common characteristic – developmental anomaly. As a result, altered growth and nutrient uptake are expressed already in early life. The placenta is a source of many proteins whose expression results in a genomic imprinting process, where some of these genes are found to be involved in the placentation process (Graves and Ranfree, 2013). IGF–II is maternally imprinted, and is normally inherited from the father, whereas the loss of IGF–II imprinting is related to an increased cell proliferation and tumour risk (Kaneda et al., 2007; Livingstone, 2013). IGFs are expressed ubiquitously in all tissues where they act in an autocrine/paracrine manner, affecting cell growth, differentiation and proliferation, mitogenesis and metabolism. Additionally, the expression of other members of the IGF system (e.g. IGFBPs, IGFBP proteases and IGF–Rs) regulates the bioavailability of these peptides and hence their numerous functions.
1.1.2. IGF binding proteins
There are six proteins belonging to the family of IGF binding proteins (IGFBP–1–6) and there is also a family of IGFBP–related proteins, that share some structural similarities with IGFBPs but bind IGFs with much lower affinity (Brahmkhatri, Prasanna and Atreya, 2015).
IGFBPs consist of 216–289 amino acids distributed in three domains (N, C, L), and it is the L domain that is not preserved between different IGFBPs, while the other two share high a degree of homology among IGFBPs and are responsible for high affinity binding of IGFs (Hwa et al., 1999). IGFBP–1, –3, and –4 bind both IGF peptides with the same affinity, while the affinity of IGFBP–2, –5 and –6 is higher for IGF–II compared to IGF–I (Firth and Baxter, 2002). IGFBPs are susceptible to different modifications: (i) IGFBP–1, –3 and –5 are phosphorylated (Jones et al., 1993; Hoeck and Mukku, 1994; Graham et al., 2007), (ii) IGFBP–3 and –4 are N–glycosylated (Firth and Baxter, 1999; Zhou et al., 2003), (iii) IGFBP–5 and –6 are O–glycosylated (Neumann, Marinaro and Bach, 1998; Graham et al., 2007).
IGF peptides have higher affinity for IGFBPs than for receptors. Hence, IGFBPs regulate the availability of peptides acting as a storage system and also serve as protective molecules as they prolong the half–life of IGFs. Though all IGFBPs form binary complexes with IGFs, some IGFBPs (e.g. IGFBP –3 and –5) also form ternary complexes. The most abundant IGFBP in the circulation is IGFBP–3, which forms ternary complexes with IGFs and the acid labile subunit (ALS). After dissociation of the 150 kDa ternary complex, IGFs form binary complexes with other binding proteins, which further transport IGFs to target tissues (Le Roith et al., 2001).
Different (patho)physiological conditions may affect post–translational modifications of IGFBPs. For example, during pregnancy, placental alkaline phosphatase, produced by the syncytiotrophoblast, dephosphorylates blood IGFBP–1, thus reducing its affinity for IGF–I (Westwood et al., 1994), resulting in the prevalence of IGFBP–1 forms with lower affinities for IGFs, further leading to higher availability of free IGF–I (Forbes and Westwood, 2008).
1.2. RECEPTORS OF THE IGF SYSTEM
As already mentioned in this chapter, due to a high degree of homology between IGFs and insulin, receptors belonging to the IGF system are not only IGF type 1 (IGF–1R) and type 2 receptor (IGF–2R), but also insulin receptor (IR) and a hybrid receptor (HyR). Their structure, function and activating cascades will be discussed in the following sections and subsections of this chapter, with an emphasis on their impact on gestation and cancer development.
1.2.1. Type 1 insulin–like growth factor receptor – IGF–1R
The IGF–1R is a 420 kDa heterotetramer, a product of the IGF–1R gene located in the 15q region. Furin cleaves the IGF–1–pro–receptor into α (135 kDa) and β (95 kDa) subunits, while two pairs of αβ heterodimers are linked via disulphide bonds (Czech and Massague, 1982; Ward et al., 2001). IGF–1R is a transmembrane protein with two α–subunits located outside of the membrane, forming an extracellular ligand–binding domain, while tyrosine kinase domains are located at the intracellular part of the protein formed by two β–subunits. Tyrosine kinase domains are located between the juxtamembrane region and the C–terminus, containing binding sites for phosphotyrosine from the signal molecules (Gatenby et al., 2013). It was shown that a heterozygous missense mutation in the IGF–1R gene can lead to severe foetal (intrauterine growth) restriction (IUGR) (Walenkamp et al., 2006), as can heterozygous mutations within the IGF–1R kinase domain (Kruis et al., 2010) or extracellular second fibronectin III domain (Wallborn et al., 2010). As high circulating levels of IGFs are found in these children, their condition is reflected by reduced IGF–1R tyrosine phosphorylation or altered cell surface expression of IGF–1R.
IGF–1R contains 16 asparagine (Asn) residues that are potential N– glycosylation sites; eleven are located in the α–subunit and five in the β–subunit (Ullrich et al., 1986). They are involved in the processing, stabilisation and localisation of the receptor (Itkonen and Mills, 2013), while the Asn913 N–glycan is found to be responsible for the IGF–1R transport to the plasma membrane (Kim et al., 2012). The importance of N–glycans is also noted at the level of sialic acid as its absence on the IGF–1R (desyalilation of the receptor) impairs cell proliferation in response to insulin (Arabkhari et al., 2010), as nicely reviewed by Ferreira et al. (2018). Under regular physiological conditions, actions of the IGFs are mediated mainly via IGF–1R.
1.2.1.1. IGF–1R cascades
Activation of the IGF–1R signalling pathway is a consequence of ligand– binding that induces conformational changes, facilitating autophosphorylation in the activation loop of the IGF–1R located in the α–subunit, leading to transphosphorylation of the opposing β–subunits (Hubbard and Till, 2000). As a result, activated IGF–1R further activates the PI3K–AKT cascade and MAPK– extracellular signal–regulated kinase (MEK)–extracellular signal–regulated kinase (ERK1/2) cascade (Coolican et al., 1997; Imai and Clemmons, 1999; Duan, Bauchat and Hsieh, 2000).
Once activated, IGF–1R interacts with adaptor proteins including Src homology collagen (SHC) proteins (p46/p52/p66) and insulin receptor substrate (IRS) (Dupont and LeRoith, 2001). SHC is primarily involved in the activation of p21ras–MAPK, which plays an important role in transduction of mitogenic signals initiated by different receptor tyrosine kinases, such as IGF–1R. After phosphorylation of IGF–1R, the receptor binds and phosphorylates SHC, leading to binding of adaptor protein growth factor–bound protein 2 (Grb2), which complexes with SOS, a p21ras guanine nucleotide exchange factor (Sasaoka et al., 2001). This action ends with ERK–1 and ERK activation, which leads to phosphorylation of cytoplasmic substrates and translocation and activation of transcription factors, enabling pro–proliferative and anti–apoptotic effects (Kolch, 2000). IGF–1R substrates are numerous: IRS, SHC, 14.3.3, CRK, CSK, PI3– kinase, SHP–2 phosphatase, Grb10 and others (Girnita et al., 2014).
IGF–1R can also utilize the components of the G–protein coupled receptor (GPCR) signalling, found to be essential for migratory and pro–survival functions controlled by IGF–I (Girnita et al., 2014). IGF–1R levels and functions are regulated by multiple post–translational modifications such as ubiquitination, sumoylation, phosphorylation and dephosphorylation (Girnita et al., 2014). Interestingly, IGF–1R propagates some completely opposing processes – growth and proliferation on one side and differentiation on the other, as well as cell adhesion on one side and motility on the other. Which process will be activated depends on the surrounding of the receptor and cell context. For example, IRS–1 is the main mediator of the mitogenic signals; however, cells that do not express IRS–1 will instead activate SHC and lead to differentiation (Romano, 2003).
1.2.2. Insulin receptor – IR
The IR is also a 420 kDa tetrameric transmembrane tyrosine kinase, encoded from the IR gene located in the 19p region. It is composed of two α– and two β– subunits. Approximately 95% of IR can be found in this heterotetrameric form, out of which 75% resides in the plasma membrane (Hwang and Frost, 1999). Due to alternative splicing, two IR isoforms exist: IR–A (a receptor isoform missing exon 11 – Ex11–, which encodes 12 amino acids from the IRs’ ectodomain at the C–terminus of α–subunit) and IR–B (a receptor isoform with preserved exon 11 – Ex11+) (Seino and Bell, 1989). The IR–A isoform is preferenatially expressed in foetal and cancer tissues, although it can also be found in the majority of other tissues (Frasca et al., 1999). IR–B is predominantly expressed during adult life, in an insulin targeted tissues, such as liver, muscle, adipose tissue and kidney. IR–A and IR–B differ in the tyrosine kinase activity and the degree of the IR’s internalisation, signal transduction and distribution in the plasma membrane depending on the membrane content of cholesterol and caveolin (Uhles et al, 2003).
IR has 18 potential N–glycosylation sites (14 on the α– and 4 on the β– subunit) out of which 16 are regularly glycosylated (Sparrow et al., 2008). The presence of sialic acid is important for IR activation after insulin binding (Dridi et al., 2013), and changes in the sialic acid content can affect receptor function (Pshezhetsky and Ashmarina, 2013).
1.2.2.1. IR cascades
Similary to IGF–1R activation, ligand binding to IR results in receptor autophosphorylation on cytoplasmic tyrosine residues and phosphorylation of the tyrosines of IRS proteins. Phosphotyrosine sites of IRS enable binding of the lipid kinase PI3K, responsible for the synthesis of phosphatidylinositol (3,4,5)– trisphosphate (PIP3) at the plasma membrane. This further activates phosphoinositide–dependent kinase that phosphorylates a threonine residue of AKT, while the serine residue of AKT is phosphorylated by mTORC2. Once activated AKT activates/phosphorylates other downstream substrates as the forkhead family box O (FOXO) transcription factors: (i) the protein tuberous sclerosis 2 (TSC2), which permits activation of mTORC1 and its downstream targets ribosomal protein S6 kinase (S6K) and sterol regulatory element binding protein 1c (SREBP1c); (ii) glycogen synthase kinase 3β (GSK3β); (iii) the RabGAP TBC1 domain family member 4 (TBC1D4). As a result of IR activation, metabolic processes are initiated as well as cell growth and differentiation. Alternative substrates of IR are Grb10, Grb14 and the suppressor of cytokine signalling (SOCS), which block IRS binding.
As can be seen for both described receptor protein kinases, phosphorylation is a crucial event for the activation or inactivation of IGF–1R and IR, e.g. tyrosine phosphorylation activates while serine/threonine phosphorylation inactivates IR and IRS proteins. These mechanisms of IR activation and inhibition of its signalling pathways are in detail and critically described in an exceptional review of Haeusler, McGraw and Accili (2018).
IR–A expression is related to a decrease in metabolic signalling of insulin and the actions of IGFs and the signalling path they trigger upon activation of IR–A. Consequent to IGF–II and proinsulin binding to IR–A, mitogenic signals are activated resulting in cell growth, proliferation and survival, being important in foetal and cancer tissues. IR–B expression, on the other hand, is associated with an increase in metabolic signalling of insulin, being important during adult life. Importantly, both IR and IGF–1R can translocate into the nucleus, thus regulating biological responses at the genomic level. This topic has been exstensively reviewed by Belfiore et al. (2017).
To increase the complexity of the IGF system, both IR and IGF–1R also have ligand–independent actions. For example, following ligand binding, due to catalytic activities of these receptors, anti–apoptotic signals are triggered enabling cell survival and resistance to apoptosis. However, unrelated to catalytic activities but only to receptors themselves, if lignads are absent and receptors are in a ligand–free form, they can support apoptosis. This effect can be reversed upon ligand binding (Belfiore et al., 2017).
1.2.2.2. IGF–1R/IR hybrid receptor – HyR
IR and IGF–1R share a high degree of similarity, ranging from 40–95% depending on the domain (Lou et al., 2006). Accordingly, one IGF–1R αβ heterodimer can dimerize with one IR αβ heterodimer (either A or B isoform) forming a hybrid receptor – HyR (Benyoucef et al., 2007). HyR can be found in tissues rich in both IR and IGF–1R, and considering there are two forms of IR, there are also two forms of HyR. Although HyR consists of the halves of two closely related receptors, there are indications that HyR is functionally much closer to IGF–1R, irrelevant to splicing, as it has rather low affinity for insulin and readily binds IGFs (Benyoucef et al., 2007).
1.2.3. Type 2 insulin–like growth factor receptor – IGF–2R
IGF–2R, also known as the cation–independent mannose–6–phosphate receptor, is a 270 kDa transmembrane protein encoded by the IGF–2R gene located in the 6q region. It mostly consists of the extracellular domain, while the transmembrane and cytoplasmic domains are much smaller (Lobel, Dahms and Kornfeld, 1988). IGF–2R is mostly expressed during foetal development (Sklar et al., 1992). Similar to other members of the IGF system, IGF–2R is also subjected to post–translational modifications. The extracellular domain contains 19 potential N–glycosylation sites, out of which at least two are glycosylated (Lobel et al., 1987). IGF–2R serves as a clearing route for IGF–II as, when bound to IGF–2R, it is directed towards lysosomal degradation (Kornfeld, 1992). There are, however, some indications of the potential signalling pathway involving IGF–2R.
1.2.3.1. IGF–2R cascades
Unlike IGF–1R and IR, IGF–2R does not contain tyrosine kinase activity or an autophosphorylation site. It is proposed that IGF–2R signalling is mediated by transactivating G protein–coupled sphingosine 1–phosphate receptors and that IGF–2R is involved in ERK1/2 activation (El–Shewy et al., 2007). Although it is thought that IGF–2R serves only for IGF–2 clearence and degradation, there are speculations that IGF–II binding to IGF–2R may be involved in mediating mitogenic effects in term placental explants (Harris et al., 2011). The exact mechanism of the potential signalling pathway mediated by IGF–2R has yet to be elucidated.
1.2.4. Physiology
Mitosis, cell growth, differentiation, migration, transformation and apoptosis are processes controlled by the IGFs. The control is exerted during embryonal development, postnatal life, maturation from childhood until adult age and ageing. As mentioned, the bioavailability and activity of IGFs are regulated by a network of IGFBPs, their proteases and IGF–Rs. In contrast to IGFs, insulin is mainly involved in metabolic processes (such as regulation of glucose concentration, protein and lipid metabolism), but mutual cross–reactivity enables IGFs to trigger metabolic responses and insulin to support cell growth (King et al., 1980). Malfunctioning in the IGF system may be associated with many pathophysiological states including cancer (Novosyadlyy and Le Roith, 2012; Nimptsch, Konigorski and Pischon, 2019).
In contrast to insulin, secreted by the pancreas, the majority of IGFs in the circulation are derived from the liver although many cells and organs can produce them locally. IGFs from the circulation exert predominantly endocrine functions, playing the role of a hormone. Peptides secreted locally exert paracrine and/or autocrine activities, expressing a role of cytokines or local growth factors. Liver is the principal organ that synthesises IGFBP–1, IGFBP–2 and IGFBP–3, but considerable quantities are produced by other organs as well, whereas synthesis of IGFBP–4, IGFBP–5 and IGFBP–6 occurs in a number of organs (Blum et al., 2018; Clemmons, 2018). IGFBPs are associated with lipid and carbohydrate metabolism, development of atherosclerosis, bone and skeletal muscle metabolism (Clemmons, 2018). Common to all IGFBPs is their capacity to control the amount of free, biologically active IGFs and, thus, to limit their presentation to cell membrane receptors. IGFBPs also protect them against proteolysis and assist in their trafficking within an organism.
IGFBPs can associate in specific complexes with other proteins beside IGFs and with other biomolecules, such as those from an extracellular matrix and glycosaminoglycans (Russo et al., 1997; Liu et al., 2014). Some IGFBPs can bind directly to cell membranes via their receptors, activating pathways other than those activated by insulin and IGFs, although they can still carry IGF ligands (Ingermann et al., 2010; Clemmons, 2018). This activity was considered as IGF– independent until recently, but a caution was drawn since some of the interactions of IGFBPs influence IGF signalling within the same cell. IGFBPs can be transported in the nucleus where they interact with nuclear proteins causing alteration of cellular physiology (Bach, 2018).
The portion of free IGFs in healthy adult persons is not greater than 1% (Juul, 2003). In contrast to insulin, stored in pancreatic granules and released upon metabolic demand, IGFs are mostly stored in blood as IGF/IGFBP complexes. The high affinity of IGFBPs for IGFs maintains the equilibrium between protein– bound and free forms of these peptides. Upon demand for IGFs, IGFBP proteases modify IGFBPs reducing their affinity and enabling take–over by membrane receptors. Some IGFBPs are said to inhibit and the others to potentiate the activity of IGFs – the difference originates from the difference in the affinity of certain IGFBP compared to the affinity of receptors for IGFs (Le Roith, 2003; Clemmons, 2018).
It is worth mentioning that the IGF system is the only one having so many specific binding proteins (beside six high–affinity, there are several low–affinity), suggesting the need for very sensitive regulating mechanisms and fine tuning in respect to the actions of IGFs (Haywood et al., 2019). Regardless of the high degree of homology between insulin and IGFs, IGFBPs do not bind insulin (Annunziata, Granata and Ghigo, 2011). Physiological concentrations of insulin in healthy adults are in the range of pM whereas the concentrations of IGF–I and IGF–II are in nM (Juul, 2003; Heinemann, 2010). Thus, since IGFs are present in 100 to 1000–fold greater concentrations than insulin, their availability must be rigorously controlled.
IGF–I is a mediator of the activity of growth hormone (GH), as GH is an up– regulator of IGF–I gene expression (Annunziata, Granata and Ghigo, 2011). In contrast to IGF–I, IGF–II is not regulated by GH (Kaplan and Cohen, 2007). Insulin/IGF signalling has been identified as one of the most important pathways that controls lifespan (Novosyadlyy and Le Roith, 2012). Blood levels of IGF–I are related to age (O’Connor et al., 1998), but they differ markedly between healthy individuals. Total IGF–I, IGFBP–3 and their ratio within one individual, however, show only small changes with age over time (Janssen, 2019). Although it is generally assumed that lower concentrations of IGF–I correlate with longevity, experimental findings do not consistently support this assumption. Lower protein intake during life may favour longevity through a process that regulates the concentration of circulating IGF–I. There may be a specific optimal age–dependent “set point” for each individual for the GH/IGF system which co– determines survival (Janssen, 2019). Enhanced signalling through the GH/IGF axis was noted to accompany an age–related disease – cancer (Anisimov and Bartke, 2013). Therefore, maintaining equilibrium within the IGF system seems to be crucial for healthy living and ageing, and mechanisms that regulate lifespan and tumour incidence are mutually linked.
IGF–II was shown to exert growth–promoting actions in placenta and it influences prenatal growth and foetal development (Cianfarani, 2012). A physiological role of this peptide, however, still remains insufficiently known. Overexpression of the IGF–II gene can result in enlarged organs and the entire body size at birth (Kadakia and Josefson, 2016). Specific polymorphisms of IGF– II have been related to an increased weight, obesity, metabolic complications and hypertension (Gaunt et al., 2001; Gu et al., 2002; Faienza et al., 2010). A degree of IGF–II methylation at birth seems to be a crucial factor for development of such changes in early childhood (Perkins et al., 2012) and it was proposed to be considered as a biomarker of intrauterine programming (Cianfarani, 2012).
IGF–1R binds IGF–I with high affinity. It also binds IGF–II and insulin but with six and a hundred fold lower affinity (Le Roith 2003; Annunziata, Granata and Ghigo, 2011). IR binds IGF–I with a hundred fold lower affinity than insulin (Ullrich et al., 1986). A hybrid IR/IGF–1R has twenty times greater affinity for IGF–I than for insulin (Sakai, Lowman and Clemmons, 2002). Tumour cells often demonstrate up–regulation or increased activity of IGF–1R (Novosyadlyy and Le Roith, 2012). As mentioned, IR exists in two isoforms where the form IR–A binds insulin and IGF–II, whereas IR–B binds predominantly insulin. The presence of specific isoforms may be associated with tumour development. Aggressive thyroid cancers, for example, overexpress IR–A, IGF–II and IGF–1R (Vella and Malaguarnera, 2018). IGF–2R binds IGF–II, but also mannose–6–phosphate (Man–6–P) and Man–6–P N–acetyl glucosamine (Nadimpalli and Amancha, 2010; Olson et al., 2014). IGF–II, after binding to IGF–2R, is most often degraded, so IGF–2R may be seen as a tumour–suppressor (Scott and Firth, 2004). Independently of the rest of the IGF system, IGF–2R acts as a lectin and regulates intracellular compartmentalisation of acid hydrolases containing Man– 6–P residues (Nadimpalli and Amancha, 2010; Olson et al., 2014).
Post–translational modifications of IGFBPs and IGF–Rs, such as glycosylation, phosphorylation, oxidation and others can influence their affinity for IGFs. Phosphorylated IGFBP–1 has high affinity for IGF–I and usually inhibits its activity. During pregnancy, however, it is dephosphorylated by placental alkaline phosphatase to generate isoforms with lower affinity and consequently, increased IGF bioavailability (Solomon et al., 2014). In foetal blood, IGFBP–1 is the principal IGFBP. Maternal diabetes is associated with reduced IGFBP–1 phosphorylation in cord serum, suggesting that diabetes–related changes may additionally increase IGF–I bioavailability and stimulate foetal growth (Loukovaara et al., 2005). Phosphorylation of Ser111 enables IGFBP–3 to induce cell apoptosis (Jafari et al., 2018). Phosphorylation and O–glycosylation of IGFBP–5 affect its binding to heparin but not to IGFs (Graham et al., 2007). Glycosylation of IGFBP–3 does not seem to influence IGF binding or formation of protein complexes, but it influences the partitioning of IGFBP–3 between the extracellular milieu and the cell surface (Firth and Baxter, 1999). Post– translational modifications of the IGFBP–binding partners also affect the formation of protein complexes. Oxidation of fibrinogen, for example, reduces its interaction with IGFBP–1 in patients with diabetes mellitus, which may be important for the duration of bleeding and the speed of wound healing (Gligorijević, Penezić and Nedić, 2017). Ageing or colon cancer caused altered glycosylation of alpha–2–macroglobulin resulting in decreased binding of IGFBP–2, increasing the amount of the free, physiologically active form of IGFBP–2 (Šunderić et al., 2019).
Both IR and IGF–1R belong to a tyrosine kinase family of receptors and they are therapeutic targets for the treatment of malignancy. Tumour cells develop resistance to targeted therapies over time by activating alternative signalling pathways. Enzymatic alteration and regulation of the N–linked glycosylation process are seen as novel targets for developing approaches to sensitize tumour cells to cytotoxic therapies (Contessa et al., 2008; de–Freitas–Junior et al., 2017). Inhibition of N–linked glycosylation of receptors in patients with congenital disorders of glycosylation (CDG) was found to impair receptor processing and surface localization (Klaver et al., 2019). Reduced fucosylation of IGF–1R, due to impaired activity of fucosyltransferase, was shown to suppress proliferation, epithelial–mesenchymal transition, migration and invasion of specific placental cells (Yu et al., 2019). Pathophysiological conditions characterised by an increased oxidative stress, such as colorectal carcinoma, may induce oxidation of IGF receptors reducing their affinity for IGFs (Nedić et al., 2013).
Growing evidence suggests that IGFBP–1 and IGFBP–2 are favourably linked with insulin sensitivity and preclinical data implicate their direct involvement in the regulation of insulin signalling and adiposity (Haywood et al., 2019). These two IGFBPs have been under investigation as therapeutic targets for obesity, metabolic disorders and diabetes. Cancer treatment, on the other hand, most often includes strategies which enable lowering of the IGF concentration and




